August 1, 2017
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Abyssomicin W
Abyssomicins are class I spirotetronate polyketides with a characteristic spirotetronate moiety. Abyssomicin C was the first reported inhibitor of the formation of p-aminobenzoate (p-ABA) from a natural source. Wang et al. [1] reported the discovery of 12 new analogs of abyssomicin as metabolites of the Streptomyces species. Amongst them Abyssomicin W (1) has an 8/6/6/6 tetracyclic core. Spectroscopic data of this compound were used to challenge ACD/Structure Elucidator Suite.
Compound 1 with a molecular formula of C20H26O8 confirmed by HR-MS has typical abyssomicin UV and NMR signatures with assumed structural differences at C-2, C-9, and C-16. Specifically, C-16 in 1 has both a unique 13C chemical shift (δC 171.5 ppm) and characteristic HMBC correlations with H-10 (δH 2.39 ppm) and H2-14 (δH 1.43, 2.45 ppm), consistent with a lack of C-2/C-16 bond and the presence of a novel lactone.
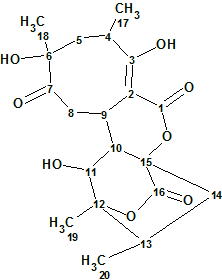
1
The NMR data from compound 1 taken from the literature are summarized in Table 1. These were entered into Structure Elucidator to automatically generate the Molecular Connectivity Diagram (MCD) shown in Figure 1.
Table 1. Abyssomicin W. NMR spectroscopic data.
| Label | dC | dC calc | XHn | dH | COSY | H to C HMBC |
| C 1 | 169.2 | 169.9 | C | |||
| C 2 | 96.6 | 97.13 | C | |||
| C 3 | 180.7 | 177.71 | C | |||
| C 4 | 30 | 32.63 | CH | 3.26 | 1.04, 1.54 |
C 5 |
| C 5 | 46.1 | 36.41 | CH2 | 1.54 | 3.26 | C 3, C 6, C 7 |
| C 6 | 78 | 78.63 | C | |||
| C 7 | 215.3 | 210.45 | C | |||
| C 8 | 43.9 | 39.66 | CH2 | 1.76 | 4.05 | C 2, C 7, C 10 |
| C 8 | 43.9 | 39.66 | CH2 | 3.29 | ||
| C 9 | 27.2 | 34.31 | CH | 4.05 | 1.76, 2.39 |
C 1, C 2, C 15 |
| C 10 | 45.3 | 39.97 | CH | 2.39 | 3.62, 4.05 |
C 2, C 14, C 15, C 16 |
| C 11 | 71.7 | 70.9 | CH | 3.62 | 2.39 | C 9, C 12, C 13 |
| C 12 | 84.9 | 86.8 | C | |||
| C 13 | 28 | 33.26 | CH | 2.44 | 0.93, 1.43 |
|
| C 14 | 40.6 | 35.94 | CH2 | 2.45 | ||
| C 14 | 40.6 | 35.94 | CH2 | 1.43 | 2.44 | C 15, C 16, C 20 |
| C 15 | 76.7 | 78.33 | C | |||
| C 16 | 171.5 | 167.75 | C | |||
| C 17 | 17.2 | 19.26 | CH3 | 1.04 | 3.26 | C 3, C 4, C 5 |
| C 18 | 28 | 27.15 | CH3 | 1.16 | C 6, C 7 |
|
| C 19 | 19 | 19.3 | CH3 | 1.28 | C 11, C 12, C 13 |
|
| C 20 | 16 | 15.67 | CH3 | 0.93 | 2.44 | C 12 |
| O 1 | OH | 13.63 | ||||
| O 2 | OH | 5.68 | C 5, C 6, C 7 |
|||
| O 3 | OH | 6.11 | C 10, C 11 |
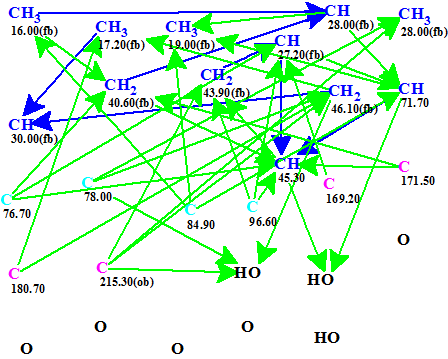
Figure 1. Abyssomicin W. Molecular Connectivity Diagram generated automatically from the data in Table 1. No manual edits were needed. The green arrows represent a max. 3 bond connectivity (HMBC data) and the blue arrows a max. 1 bond connectivity (COSY data).
No edits of MCD were made. Discrete Structure generation was initiated using the software default parameters. Structure generation gave the following result: k→1424→41→41, tg = 1.4 s
The six top ranked generated structures are shown in Figure 2. The structures are ranked according to lowest mean deviation of experimental from HOSE-codes predicted 13C chemical shift difference.
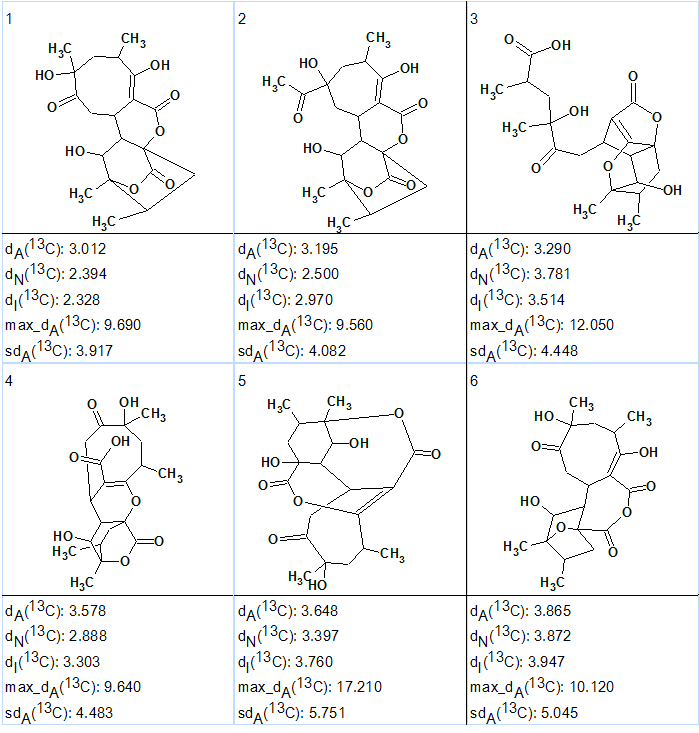
Figure 2. Abyssomicin W. Six top ranked structures of the output file.
Structure #1 coincides with the structure of Abyssomicin already reported [1].
As the difference between deviations calculated for structures 1 and 2 is small some further confirmation is needed. This could be achieved by either additional NMR data (NOESY/ROESY or additional HMBCs at different coupling constant settings) or by DFT-based chemical shift prediction [2,3].
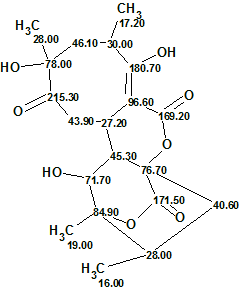
2
References:
- X. Wang, S. I. Elshahawi,W. Cai,Y. Zhang, L. V. Ponomareva, X. Chen, G. C. Copley, J. C. Hower, C.-G. Zhan, S. Parkin, J. Rohr, S.G. Van Lanen, K. A. Shaaban, J. S. Thorson. (2017). Bi- and Tetracyclic Spirotetronates from the Coal Mine Fire Isolate Streptomyces sp. LC-6-2. J. Nat. Prod., 80: 1141−1149.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.
- A. V. Buevich, M. E. Elyashberg. (2016). Synergistic combination of CASE algorithms and DFT chemical shift predictions: a powerful approach for structure elucidation, verification and revision. J. Nat. Prod., 79 (12): 3105–3116.


