March 1, 2019
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
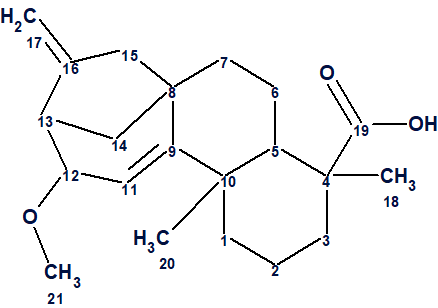
Ent-Kaurane-Type Diterpenoid
The majority of the Asteraceae family of plants are herbaceous, with trees and shrubs being rare. They can be found all over the world, with the exception of Antarctica. Aspilia belong to this family and is known to have biological activity. This is attributed to the presence of kaurane-type diterpenoids and sesquiterpene lactones. In fact Aspilia pluriseta Schweinf has been used in traditional medicine. This plant is commonly known as “Dwarf Asprilia” and can be found in Kenya.
Yaouba et al [1] reported the isolation and phytochemical investigation of the constituents of Aspilia pluriseta Schweinf and Aspilia mossambicensis. The authors [1] isolated, identified and comprehensively investigated compound 1.
1
Compound 1 was isolated as colorless crystals (m.p. 184–186 °C) from the CH2Cl2/MeOH (1:1) extract of the roots of Aspilia pluriseta. HRMS showed a [M–H]− ion peak at m/z = 329.2191, which is in agreement with the molecular formula C21H30O3. The structure of compound 1 was elucidated by authors [1] using 1D and 2D NMR (HSQC, COSY, HMBC, NOESY) data.
These data (Table 1) were used to test ACD/Structure Elucidator.
Table 1. Spectroscopic 1D and 2D NMR data of compound 1
| Label | δC | δC calc a | CHn | δH | M(J) | COSY | H to C HMBC |
| C 1 | 40.6 | 41.28 | CH2 | 1.9 | u | C 2, C 20, C 3, C 10 | |
| C 1 | 40.6 | 41.28 | CH2 | 1.14 | u | 1.43 | C 8 |
| C 2 | 20 | 19.89 | CH2 | 1.79 | u | ||
| C 2 | 20 | 19.89 | CH2 | 1.43 | u | 0.93, 1.14 | C 3, C 10, C 1, C 4, C 5 |
| C 3 | 38.1 | 38.2 | CH2 | 2.08 | u | ||
| C 3 | 38.1 | 38.2 | CH2 | 0.93 | u | 1.43 | C 2, C 18, C 1, C 4, C 19 |
| C 4 | 44.6 | 45.39 | C | ||||
| C 5 | 46.1 | 48.37 | CH | 1.56 | u | C 20, C 18, C 7, C 10, C 4, C 9, C 19 | |
| C 6 | 18.3 | 19.78 | CH2 | 2.43 | u | C 8 | |
| C 6 | 18.3 | 19.78 | CH2 | 1.82 | u | 1.42 | C 7, C 3, C 10, C 4, C 5 |
| C 7 | 28.9 | 29.89 | CH2 | 1.95 | u | ||
| C 7 | 28.9 | 29.89 | CH2 | 1.42 | u | 1.82 | C 6, C 8, C 5, C 15, C 9 |
| C 8 | 43.4 | 44.62 | C | ||||
| C 9 | 160.2 | 157.3 | C | ||||
| C 10 | 38.9 | 39.29 | C | ||||
| C 11 | 115.3 | 117.45 | CH | 5.3 | u | 3.38 | C 20, C 10, C 8, C 13, C 15, C 12, C 9 |
| C 12 | 81.7 | 79.85 | CH | 3.38 | u | 2.89, 5.30 | C 20, C 13, C 21, C 11, C 16, C 9 |
| C 13 | 43.7 | 48.69 | CH | 2.89 | u | 1.31, 3.38 | C 10, C 15, C 12, C 11, C 16 |
| C 14 | 40.5 | 42.42 | CH2 | 1.58 | u | ||
| C 14 | 40.5 | 42.42 | CH2 | 1.31 | u | 2.89 | C 7, C 8, C 13, C 15, C 12, C 16, C 9 |
| C 15 | 47.1 | 48.15 | CH2 | 2.08 | u | C 7, C 8, C 17, C 16, C 9 | |
| C 15 | 47.1 | 48.15 | CH2 | 2.35 | u | C 7, C 14, C 13, C 17, C 16, C 9 | |
| C 16 | 152.9 | 150.24 | C | ||||
| C 17 | 108.1 | 106.44 | CH2 | 4.94 | u | ||
| C 17 | 108.1 | 106.44 | CH2 | 4.84 | u | C 13, C 15, C 12, C 16 | |
| C 18 | 28.2 | 24.38 | CH3 | 1.17 | 0 | C 3, C 8, C 4, C 5, C 19 | |
| C 19 | 183.2 | 182.56 | C | ||||
| C 20 | 23.4 | 26.11 | CH3 | 1.01 | 0 | C 10, C 1, C 5, C 9 | |
| C 21 | 56.5 | 55.5 | CH3 | 3.34 | 0 | C 12 |
a 13C chemical shift prediction was performed by the HOSE code-based algorithm implemented into ACD/Structure Elucidator.
A Molecular Connectivity Diagram (MCD) automatically created by the program is shown in Figure 1.
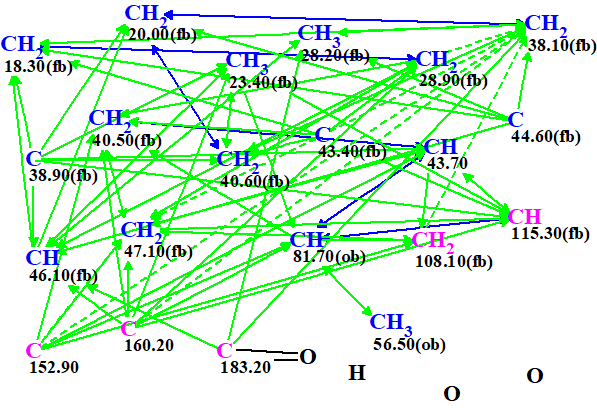
Figure 1. Slightly edited molecular connectivity diagram.
MCD overview. The hybridization of carbon atom CH 115.3 was automatically assigned by the program as not sp (sp2 or sp3), and this atom was colored in light blue. Because carbon C 183.2 is certainly a carbonyl this atom was manually connected to an oxygen with a double bond. Carbon atom 81.70 (ob) has to be connected with O-CH3 (56.5) or with OH, therefore atom CH 115.3 can not be a central atom of the fragment O-CH-O. With this in mind, hybridization of this atom was assigned as sp2 (colored by violet). The two mentioned edits were done to the initial MCD.
The MCD checking for presence of non-standard correlations (NSC) was completed with the following program message: “The minimum number of NSCs is 5”. Hence the program concluded that, on the basis of data logical analysis, collective HMBC and COSY data contained at least five long-ranged correlations whose lengths is larger than 3 (nJCH,HH, n>3).
Therefore, Fuzzy Structure Generation [2,3] was initiated with options that were set by the program automatically. These options assume the presence of unknown number of NSCs having unknown lengths.
Results: k = 1, tg = 3 min. During the structure generation, the program detected 8 NSCs and checked 1,316,554 from 1,420, 494,075 theoretically possible connectivity combinations. This means that 1,316,554 attempts to generate structures from a given current connectivity combination were made. The single generated structure along with its average deviations calculated by three empirical methods of 13C chemical shift prediction (HOSE code-based, dA; neural networks algorithm, dN; incremental approach, dI) is shown in Figure 2.
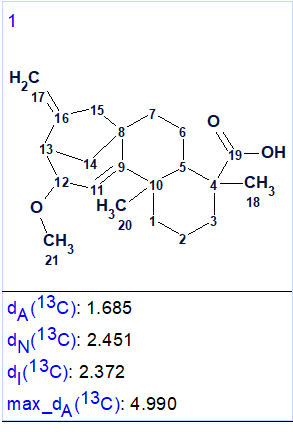
Figure 2. Output structure.
We see that the generated structure coincides with the structure of compound 1 as determined in [1]. A small value of all the average deviations supports the validity of the structure elucidated with ACD/Structure Elucidator. The high accuracy of 13C chemical shift prediction, when using HOSE code-based algorithm, is illustrated by the “colored” structure where a color labels deviation values calculated for each carbon atom:
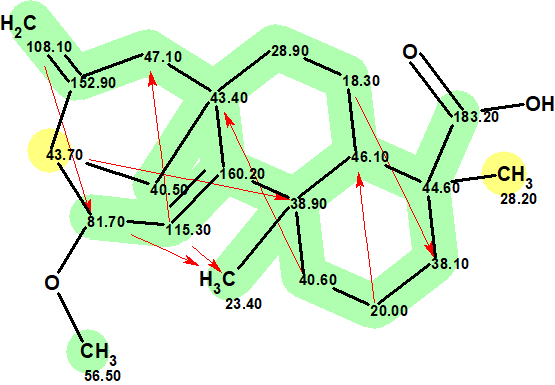
Deviations whose value is less than 3 ppm are marked by green color, while the ones between 3 and 15 ppm marked with yellow. The red arrows show the 8 NSCs detected by the program during the process of fuzzy structure generation. The connectivity C43.70/C 38.90 is two chemical bonds longer in comparison to the length of a “standard connectivity”, while other NSCs are of the n=4 type (nJCH). Even if the C43.70/C 38.90 connectivity is not real and is a consequence of an artifact in the HMBC pattern, its presence did not prevent the program to elucidate a right structure.
References
- S. Yaouba, A. Valkonen, P. Coghi, J. Gao, E. M. Guantai, S. Derese, V. K. W. Wong, M. Erdélyi, A. Yenesew. Crystal Structures and Cytotoxicity of ent-Kaurane-Type Diterpenoids from Two Aspilia Species. Molecules, 2018, 23, 3199; doi:10.3390/molecules23123199
- Elyashberg, M. E.; Williams, A. J.; Blinov, K. A. Contemporary Computer-Assisted Approaches to Molecular Structure Elucidation; RSC, Cambridge, 2012.
- M. E. Elyashberg, A. J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data. The Art of Solving Problems. Springer.


