February 1, 2021
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Peyssonnoside A
Diterpenoids and their analogues and derivatives are well known and highly varied compounds. Cyclopropane groups are also frequently encountered in natural products, but it is rare that highly substituted and/or embedded cyclopropanes are reported. The structural novelty of such compounds together with their bioactivity are valuable for medicinal, synthetic and biological chemists.
Peyssonnoside A (1), an unusual diterpene glycoside with a sterically hindered cyclopropane motif, was isolated by Kubanek and co-workers [1] from the mid-polarity fraction of the Peyssonnelia sp. extract.
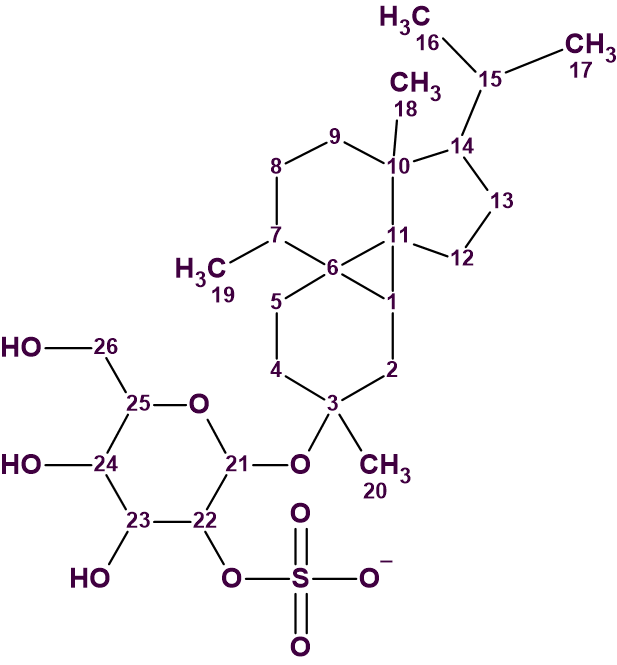
1
Its planar structure, relative and absolute configuration were determined by spectroscopic methods and were sequentially supported with computer-assisted structure elucidation (CASE), density functional theory (DFT)-based prediction of 13C chemical shifts and simulation of experimental ORD data.
HRESIMS data of 1 (m/z 531.2625 [M]−) indicated a molecular formula of C26H43O9S.
The 1D and 2D NMR data used for the planar structure elucidation are shown in Table 1.
Table 1. NMR spectroscopic data.
| Label | δC | δCcalc (HOSE) | XHn | δH | COSY | H to C HMBC |
| C 1 | 13.900 | 14.090 | CH | 0.690 | 0.85, 2.16 | C 5, C 6, C 12, C 7, C 11, C 10, C 3 |
| C 2 | 29.800 | 35.270 | CH2 | 0.850 | 0.69 | C 1, C 11 |
| C 2 | 29.800 | 35.270 | CH2 | 2.160 | 0.69, 1.35 | C 1, C 4, C 11, C 3 |
| C 3 | 72.500 | 77.200 | C | |||
| C 4 | 33.900 | 35.060 | CH2 | 1.350 | 0.75, 1.02, 1.16, 1.96, 2.16 | C 1, C 19, C 5, C 6, C 8, C 2, C 9, C 3 |
| C 4 | 33.900 | 35.060 | CH2 | 0.770 | 1.15, 1.96 | C 5, C 20, C 2, C 21 |
| C 5 | 21.300 | 22.610 | CH2 | 1.150 | 0.77 | C 1, C 6, C 4, C 11, C 3 |
| C 5 | 21.300 | 22.610 | CH2 | 1.960 | 0.77, 1.35 | C 6, C 4, C 7, C 3 |
| C 6 | 24.200 | 26.500 | C | |||
| C 7 | 35.000 | 31.520 | CH | 1.350 | ||
| C 8 | 26.800 | 28.410 | CH2 | 1.160 | 1.35, 1.47 | C 10 |
| C 8 | 26.800 | 28.410 | CH2 | 0.750 | 0.91, 1.35 | C 19, C 6, C 7, C 9, C 10 |
| C 9 | 39.200 | 35.420 | CH2 | 1.470 | 1.16 | C 18, C 8, C 7, C 11, C 10 |
| C 9 | 39.200 | 35.420 | CH2 | 0.910 | ||
| C 10 | 41.600 | 43.200 | C | |||
| C 11 | 36.200 | 39.220 | C | |||
| C 12 | 25.100 | 27.420 | CH2 | 1.270 | 1.19, 1.73 | C 1, C 6, C 13, C 11, C 10, C 14 |
| C 13 | 27.900 | 27.860 | CH2 | 1.190 | 1.27, 1.56, 1.73 | C 18, C 12, C 15, C 10, C 14 |
| C 13 | 27.900 | 27.860 | CH2 | 1.730 | 1.19, 1.27 | C 18, C 12, C 15, C 11, C 10, C 14 |
| C 14 | 61.400 | 53.820 | CH | 1.190 | ||
| C 15 | 28.500 | 29.980 | CH | 1.560 | 0.87, 0.91, 1.19 | C 17, C 16, C 13, C 10, C 14 |
| C 16 | 23.300 | 23.260 | CH3 | 0.910 | 0.75, 1.56 | C 18, C 17, C 15, C 7, C 10, C 14 |
| C 17 | 23.000 | 21.850 | CH3 | 0.870 | 1.56 | C 16, C 15, C 14 |
| C 18 | 19.000 | 20.710 | CH3 | 0.640 | C 1, C 11, C 9, C 10, C 14 | |
| C 19 | 19.100 | 18.680 | CH3 | 1.020 | 1.35 | C 6, C 8, C 7 |
| C 20 | 25.800 | 30.220 | CH3 | 1.000 | C 1, C 5, C 2, C 4, C 3 | |
| C 21 | 94.700 | 95.170 | CH | 4.390 | 3.65 | C 3, C 25, C 23, C 22 |
| C 22 | 79.300 | 79.320 | CH | 3.650 | 3.44, 4.39 | C 23, C 21 |
| C 23 | 76.700 | 77.990 | CH | 3.440 | 3.00, 3.65 | C 24, C 25, C 22, C 21 |
| C 24 | 70.700 | 71.690 | CH | 3.000 | 3.13, 3.44 | C 26, C 25, C 23 |
| C 25 | 76.100 | 77.290 | CH | 3.130 | 3.00, 3.37, 3.72 | C 26, C 24, C 23, C 21 |
| C 26 | 61.800 | 62.430 | CH2 | 3.370 | 3.13 | C 24, C 25 |
| C 26 | 61.800 | 62.430 | CH2 | 3.720 | 3.13 | C 24, C 25 |
| O 1 | OH | 5.630 | C 24, C 23, C 22 | |||
| O 2 | OH | 4.970 | C 24, C 25, C 23 | |||
| O 3 | OH | 4.220 | C 26, C 25 |
The data presented in Table 1 were entered into the ACD/Structure Elucidator Expert System to challenge the software. The Molecular Connectivity Diagram (MCD) automatically created by the program is shown in Figure 1.
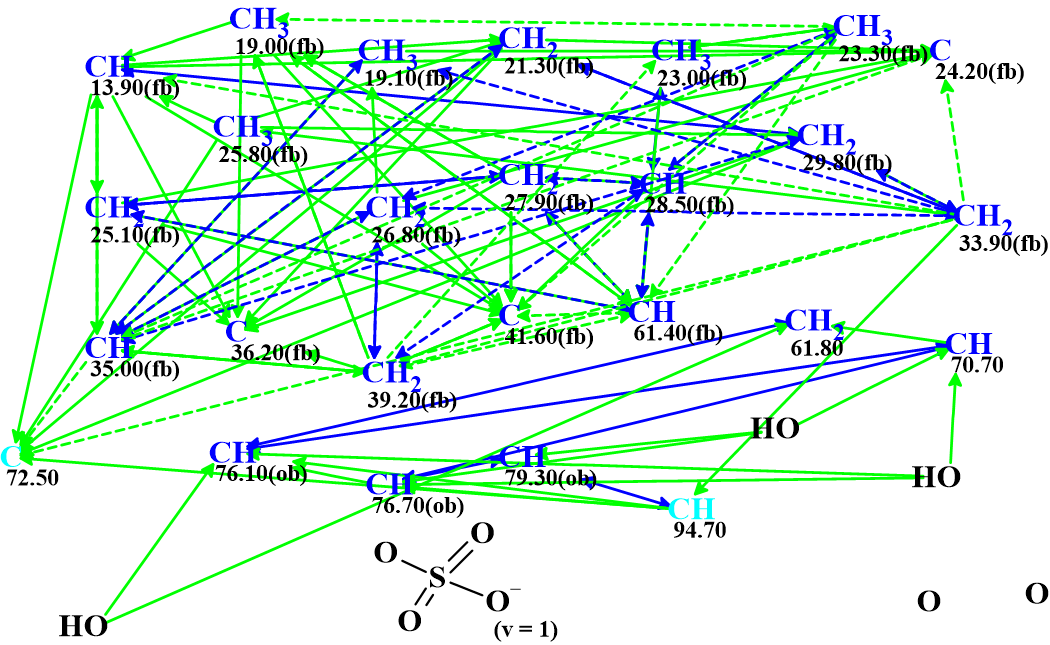
Figure 1. Molecular connectivity diagram.
MCD overview: All carbon atoms colored in blue color have sp3 hybridization. Light blue carbons C 72.50 and C 94.7 are classified by the program as not sp (sp2 or sp3). Atoms for which neighboring with heteroatoms is forbidden or obligatory are marked by labels “fb” or “ob” correspondingly. The HMBC correlations are marked by green arrows, while those derived from COSY by blue. Dotted line arrows show ambiguous correlations. The latter appear due to the presence of three pairs of identical 1H chemical shifts in the NMR data (see the colored 1H chemical shifts in Table 1). All data were defined by the program automatically. The sulfate moiety which was deduced by authors was manually drawn. The presence of this group could probably be confirmed by and IR spectrum, but unfortunately such data were not present in the article [1].
The data presented in the MCD was checked for the presence of contradictions, that is correlations whose length is non-standard (nJCH, HH, n>3). These correlations are referred to as NSCs (Non-Standard Correlations). The MCD checking was completed by the following program message: “The minimum number of non-standard connectivities is 3”.
Therefore “Fuzzy Structure Generation” (FSG) was initiated. This procedure was designed to allow structure generation to proceed in the presence of an unknown number of NSCs whose lengths are also unknown. FSG was carried out with the automatically set options.
Result: k=1, generation time tg = 2 m 25 s, and 6 NSCs (from a total of 90 connectivities) have been detected and extended during generation.
13C chemical shift prediction was performed afterwards, using the three methods available in ACD/SE: HOSE-code based approach, Neural Networks and Incremental method. The chemical shift average deviations dA, dN and dI were also calculated (designations correspond to the above-mentioned methods). The solution to the problem is shown in Figure 2.
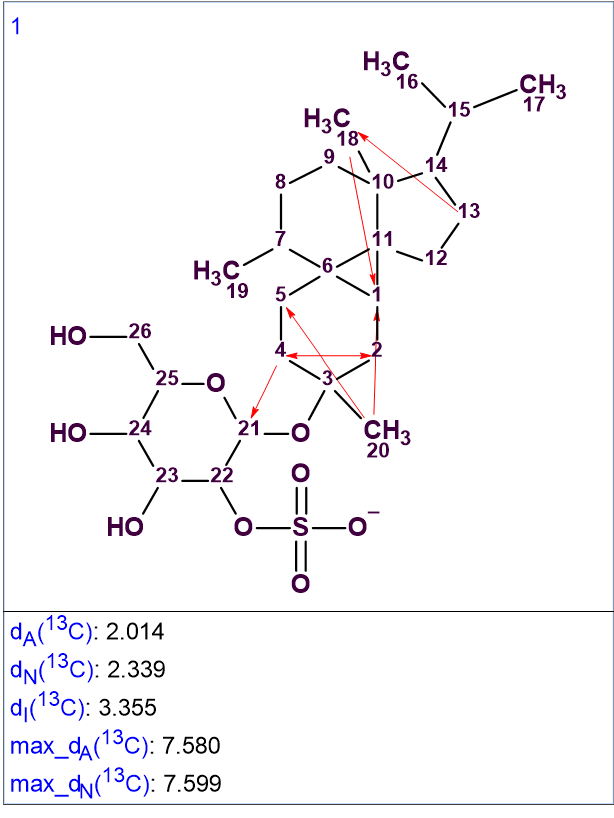
Figure 2. The single generated structure.
The solution obtained is identical to the structure 1 determined by the authors [1]. Five one-sided red arrows denote NSCs determined in HMBC data, and one double-side arrow (2-4) indicates an NSC in COSY data. Note that the solution to the problem is unique despite of the presence of six NSCs. This is due to the large number of cross-peaks detected in HMBC and COSY spectra, which is illustrated in Figure 3, where all 2D NMR connectivities detected are shown on the structure.
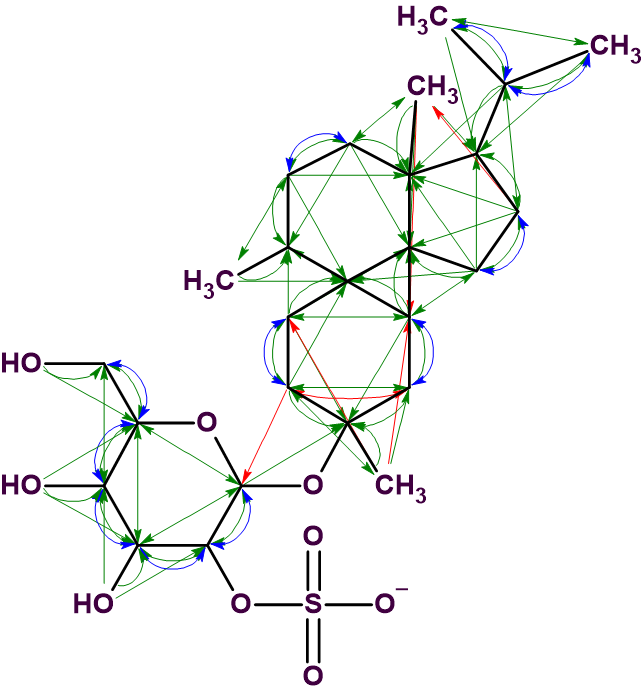
Figure 3. Structure 1 with all connectivities visualized.
Interestingly the authors [1] state that the CASE system they used (not ACD/SE) generated more structures that were subsequently ruled out with additional NMR data. It appears that these additional structures were generated as the authors used the CASE system to generate sub-structures without the sugar ring which they then evaluated manually (Figure 4), and not the whole structure in one go. It is clear that these additional structures contradict the data even from the 1D spectrum, as seen in the captions in Figure 4, so they should not have been generated at all. Unfortunately, these structures are shown without the assignment they were given so it is not possible to understand what exactly happened and see how and if the CASE system used dealt with the large number of NSCs.
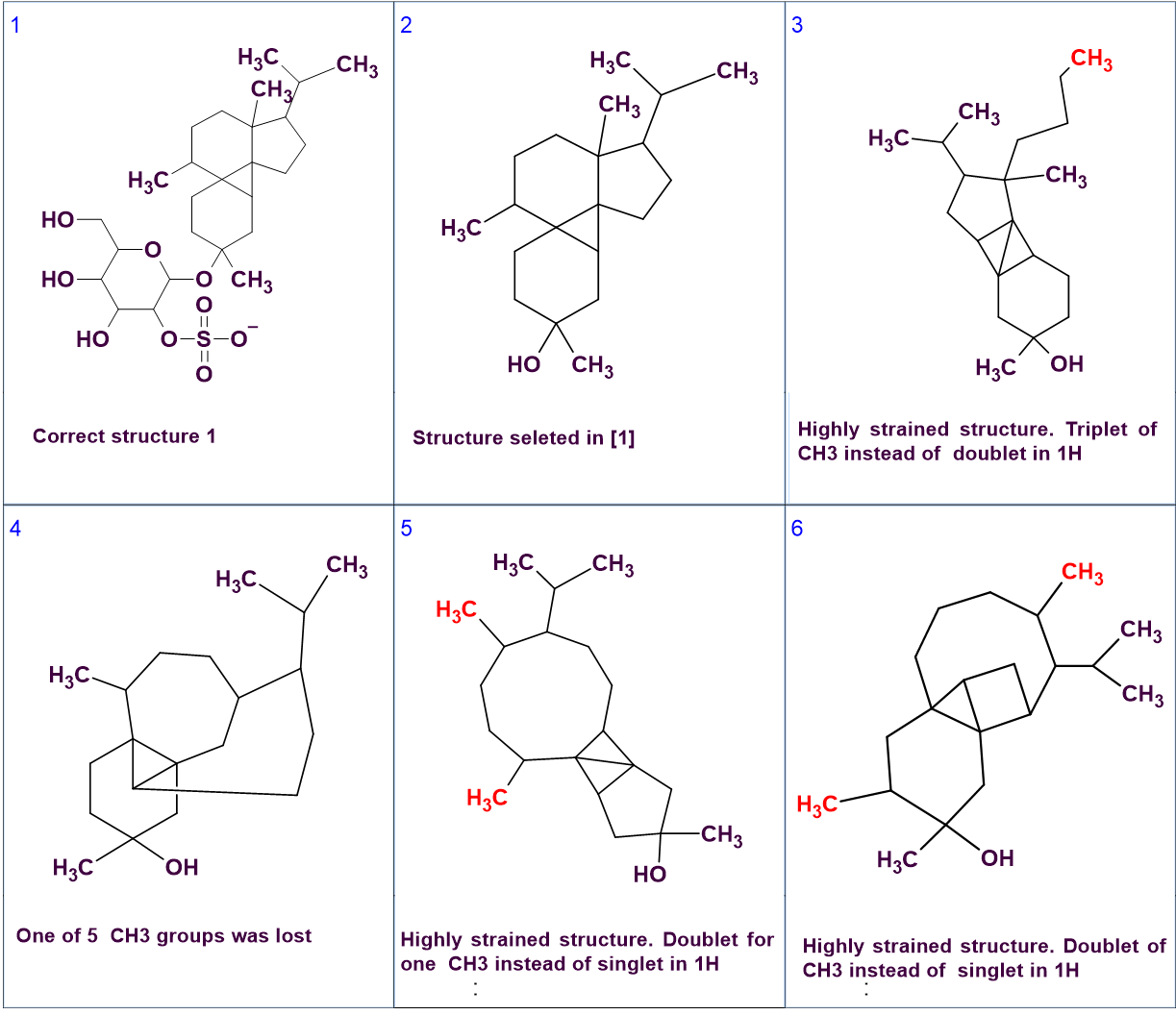
Figure 4. Correct structure #1 and candidate structures produced by the CASE system used by the authors [1].
The structure of peyssonnoside A with carbon atoms automatically assigned is shown below.
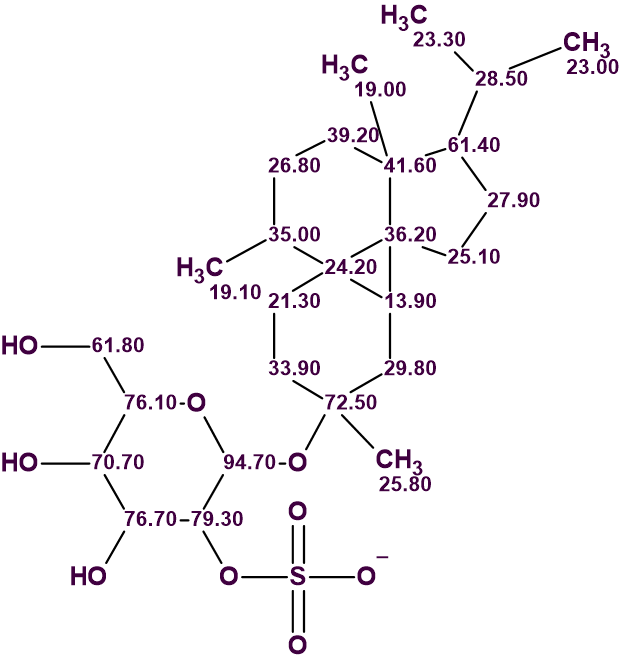
In conclusion, the unusual structure of compound 1 whose 2D NMR data contained 6 NSCs was unambiguously elucidated using ACD/Structure Elucidator in one go, in a fully automatic mode without the need for extensive editing of the MCD or breaking down the problem to smaller pieces.
References:
- B. K. Chhetri, S. Lavoie, A. M. Sweeney-Jones, N. Mojib, V. Raghavan, K. Gagaring, B. Dale, C. W. McNamara, K. Soapi, C. L. Quave, P. L. Polavarapu, J. Kubanek. (2019). Peyssonnosides A−B, Unusual Diterpene Glycosides with a Sterically Encumbered Cyclopropane Motif: Structure Elucidation Using an Integrated Spectroscopic and Computational Workflow. J. Org. Chem., 84: 8531−8541.


