New Features and Updates in V2025
Version 2025 of Structure Elucidator Suite adds new features including the ability to import data from NMR eData SD files and automatic detection of para-disubstituted phenyl fragments. There are also improvements to handling HMBC data of methyl-containing structures for faster elucidation. Read below for details, and contact us for help upgrading your software.
Highlighted Features | Improved Features | Data Import & Export | Other Techniques
Highlighted Features
Reduce Structure Generation Time for Structures Containing Para-Disubstituted Phenyl Fragments
- You can now check the dataset for the presence of one or more 1,4-disubstituted phenyl fragments during the general auto-MCD check procedure. Identifying fragments prior to structure generation reduces the scale of this computational task and can significantly reduce the amount of computation required.
Accelerate Elucidations of Methyl-Containing Structures with Improved HMBC Bond Length Validation
- You can now elucidate structures containing methyl groups in a fraction of the time with an improved algorithm that better handles their corresponding HMBC signals. This allows for more representative bond length estimation of all HMBC peaks, leading to better constraints and faster elucidation.
Identify Atoms Corresponding to Row in User Tables
- You can now easily identify the atom in the proposed structure or User Molecular Connectivity Diagram (UMCD) corresponding to the selected row in a User Table, as it is now automatically indicated.
Improved Features
Data Analysis & Processing
Display Positive and Negative 1D Projections of Phase-Sensitive 2D Methods by Default
- You can now see both positive and negative 1D projections when displaying predicted 2D spectra involving phase-sensitive methods (i.e., HSQC-DEPT)
Predict Spectra in D2O and Deuterated Methanol that Better Reflect Experimental Results
- You now have the option to predict spectra in D2O and deuterated methanol that ignore the exchangeable protons of the structure not observed experimentally due to exchange with deuterium from the solvent
Allow for Small Variations in the Integral Range of Peaks in External Standard qNMR
- You can now do external standard qNMR calculations using peaks whose integral ranges are not identical between replicates, which often occurs when working with manually defined integrals
New Parameters to Support Broader Workflows in External Standard qNMR
- You can now perform external standard qNMR calculations using replicate samples with different masses but constant volume with the help of:
- New parameters added to Concentration Calculation Tool and default report template
- Average w/w% (averaged across different replicates)
- w/w% RSD%
- Maximum Peak-to-Peak RSD% (intrasample)
- Mass and volume information included in the table of the Used Replicates popup menu
- New parameters added to Concentration Calculation Tool and default report template
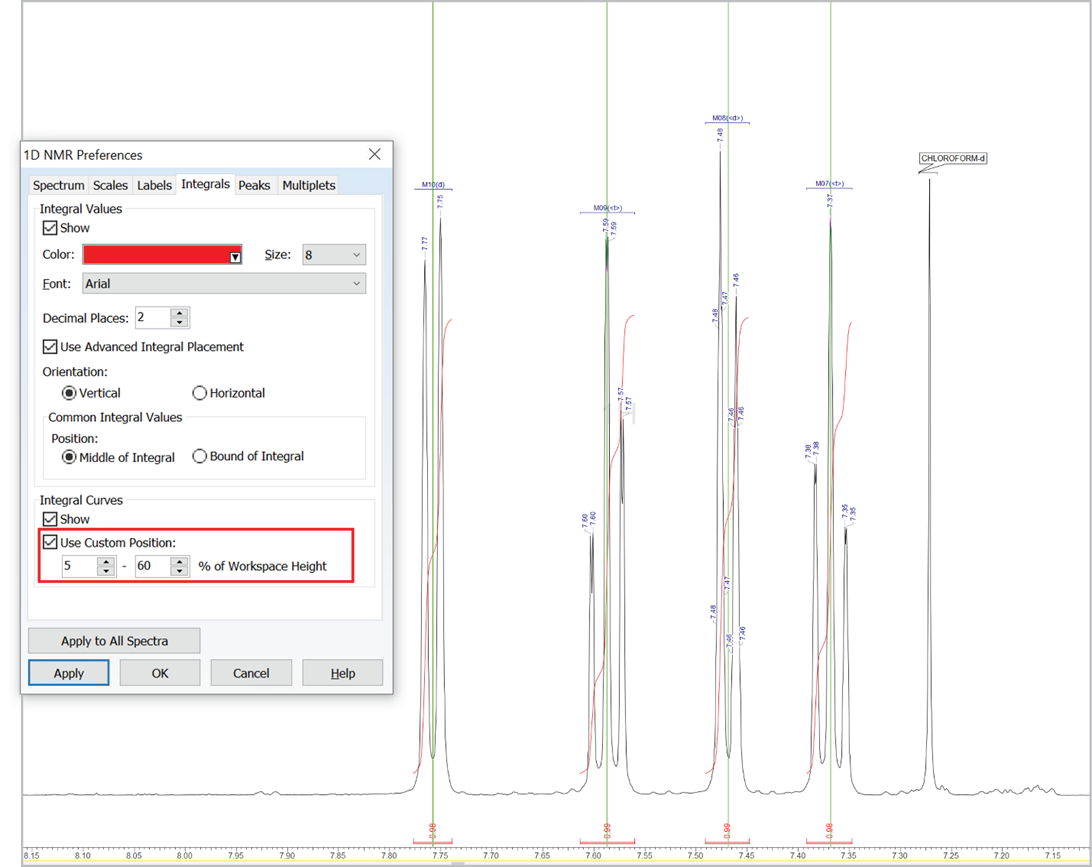
Customize the Position of Integral Curves
- You can now set a custom position for integral curves in the processing interface and reports by defining the desired vertical span of the highest curve in the displayed spectral region (as % relative to height of spectrum window, regardless of the vertical scale factor value)
Apply Unique Baseline Correction Parameters in Each Dimension of a 2D NMR Spectrum
- You can now apply baseline correction in each dimension of a 2D spectrum independently, allowing you to apply different parameters in each
Automatically Analyze Multiplets in 19F NMR Spectra
- You can now automatically analyze 19F spectra of structures with multiple fluorine atoms with an improved algorithm for better detection of first-order multiplet patterns
Improved Identification of Peaks Near Water in 1H NMR Spectra in DMSO
- You can now more accurately identify spectral peaks that appear at a similar position as the water peak in 1H spectra in deuterated DMSO (e.g., N-Me, O-Me, etc.) with an improved automatic analysis algorithm
User-Added Solvents/References Available to Macro Commands for NMR
- You can now utilize user-added solvents or references in macro commands by first invoking the new LoadSolventsTable macro command
Databasing
Audit Trail & Record History Improvements
- You can now add a column in the audit trail table to display the signature status of a record for quick identification of signed/unsigned entries
- You can now see the full history of record signing/un-signing in the Records History dialog box
Reporting
Report Only Data from the Current Spectrum in the Table of Spectral Data
- You now have the option to include only data from the current spectrum in the Table of Spectral Data of a report
Optimize the Display of NMR Spectra in Reports
- You can now easily remove whitespace at the outer edges of frequency axes in 1D and 2D NMR spectra in reports. The new Auto Fit option in the Report Page Setup dialog box automatically selects a display region within ±0.5 ppm (¹H) or ±5 ppm (other nuclei) of the outermost peaks.
Summarize w/w% Values in qNMR Reports
- You can now include w/w% values for each replicate in the Sample Data table of a qNMR report
Ease of Use
- You can automatically revert back after showing all tabs of open data with the new Fit to Tab Names button
- You can now quickly select/unselect all NMR integrals except for that of the solvent in external standard qNMR with new a context menu
Data Import & Export
Import Data from NMR eData SD Files
- You can now import peak picking, structure, and assignment information into User Tables for use in structure generation
Import Analyzed NMR Spectra from Multispectra JCAMP Files
- You can now import processed and assigned 1D and 2D NMR spectra from multispectra JCAMP files produced by third-party software as multiple separate spectra
New Formats for MS & Chromatography Data Import
- You can now import data from the following formats:
- TetraScience for Empower (*.json) DAD (LC-UV)
- Waters Micromass Fraction Lynx (*.fpt)
Improved Import Filters for MS & Chromatography Data
- You can expect improved import filters for easier data analysis:
- Shimadzu IO module
- Waters Micromass OpenLynx RPT data to include labels of ELSD traces
- Agilent Chemstation (*.D) folders containing multiple *.ch files will be imported together
Expanded and Improved Export of Data from Database Records
- You can now export all record components as *.csv and *.txt files
- Options for exporting structure data to SD files have been expanded to include the MOL file version, extensions, and labels
Features for Other Techniques
As always, you can process data from other analytical techniques in Structure Elucidator Suite. We’ve improved features for these techniques as well:
Processing MS & Chrom Data with Structure Elucidator Suite
Data Analysis & Processing
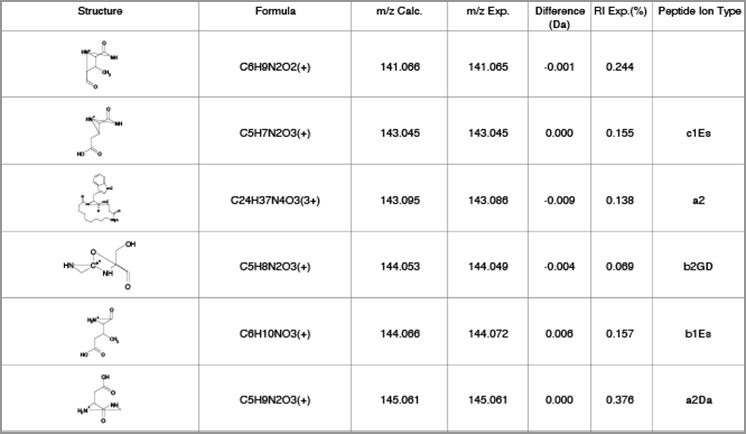
Improved Peptide Fragmentation
You can now more easily interpret your spectra for complex linear and cyclic peptides.
- Fragment and label complex linear peptides as a, b, c, x, y, and z ions
- Generate fragment ions for cyclic peptides based on the Roepstorff-Fohlman-Biemann nomenclature
- Cyclic fragments are labelled using the anJK format where:
- a = type of peptide ion (e.g., a, b, x, or y)
- n = number of amino acid residues within the fragment
- J = residue at the N-terminus end of the cycle break
- K = residue at the opposite side of the cycle break
- Cyclic fragments are labelled using the anJK format where:
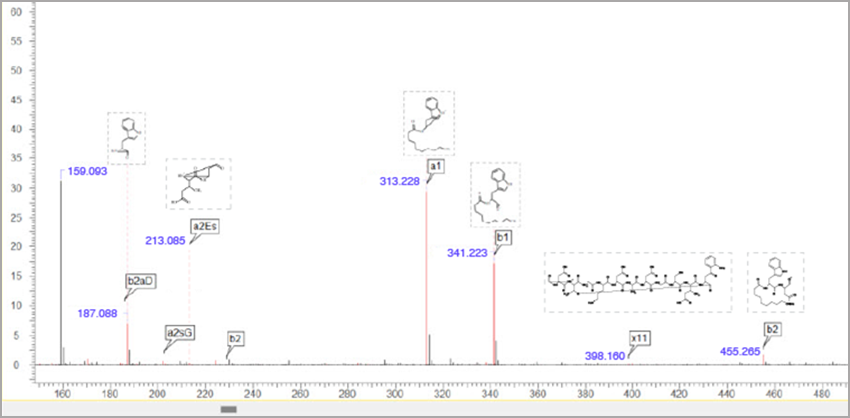
Mass spectrum and fragmentation table showing fragmentation of cyclic peptide and anJK peptide ion type.
Simplified User Input and Analysis
Improvements simplifying user input and analysis in quantitation have been made:
- Calculate and display the mean and %RSD for retention time and area in the calibration table for replicates
- You can now transfer the standard compound name to Table of Peaks
Better Efficiency in Intelligent Component Recognition Workflow (IXCR)
- You can use a dynamic mass range to reduce the spectral search criteria to 0.5 Da above the molecular ion which will also change your HQI score accordingly
- Automatically populate desired indices into the Table of Components when they exist in a searched database to more easily compare retention characteristics
Enhanced Data Analysis and Interpretation
Several improvements have been made to the IX, IXCR, and ITA algorithms, including:
- You can now control the conversion of spectra from a profile to centroid spectra (IX 2.0, IXCR)
- You can now interrogate your data further with automatic creation and display of MS3 Component spectra following processing (manual assignment, IX 2.0, IXCR, and ITA)
Simplified Chromatogram Peak Detection
- Filter your picked peaks by setting a minimum area threshold (as a percentage of the total) and automatically integrate all peaks above this limit
Improved Visualization of Chromatogram Structures
- You can adjust the size of structures displayed on peaks to better visualize those of interest
Reporting
Display Intensity Values on MS Peaks
- You can now view detector and relative intensity values directly over mass spectral peaks for easier analysis
Improved Visualization of Structures on Chromatograms
- You now have the ability to display chemical structures more clearly on chromatographic peaks in your reports
Updated Default MS Reports
- You now have more control over MS reports with the ability to generate improved default mass spectra reports
Expanded Component Reporting Options
- You can now report component mass spectra by collision energy, and include component UV spectra, giving you a more complete view of your data
Ease of Use
- You can now remove any unnamed components from the Table of Components with a single click
- You can now more easily set modifications to your mass spectra analysis with an improved Component Interpretation Options dialog box
- You can now display intensity values (detector or relative) over mass spectral peaks to more easily analyze your data
Want to learn more?
Read more about the full features of Structure Elucidator Suite, or contact us for help upgrading your software.