December 1, 2025
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Computer-Assisted Elucidation of Spiromastixone Z4 Using Structure Elucidator Suite
Halogenated natural products play a significant role in drug discovery and development. Approximately 27% of small-molecule pharmaceuticals and over 80% of agrochemicals contain halogen atoms. Owing to the high levels of chloride and bromide ions in seawater, marine natural products serve as a major reservoir of halogenated compounds. Because chloride is present at concentrations nearly three orders of magnitude higher than bromide, chlorinated metabolites occur in the ocean at roughly nine times the frequency of brominated ones. The type of halogen incorporated can also influence the biological activity of these compounds.
During a search for bioactive secondary metabolites from the deep-sea-derived fungus Spiromastix sp. MCCC 3A00308, Lin and co-workers characterized a series of chlorinated compounds, including the antibacterial depsidones spiromastixones A–S. To investigate the effect of bromination on the antibacterial activities of spiromastixones, the same group fermented the fungus Spiromastix sp. MCCC 3A00308 using rice media supplemented with sodium bromide. As a result, eleven new brominated depsidones, spiromastixones U-Z5 were isolated and identified using MS and NMR methods [1].
One of the compounds included in this series was brominated depsidone spiromastixone Z4 (1).
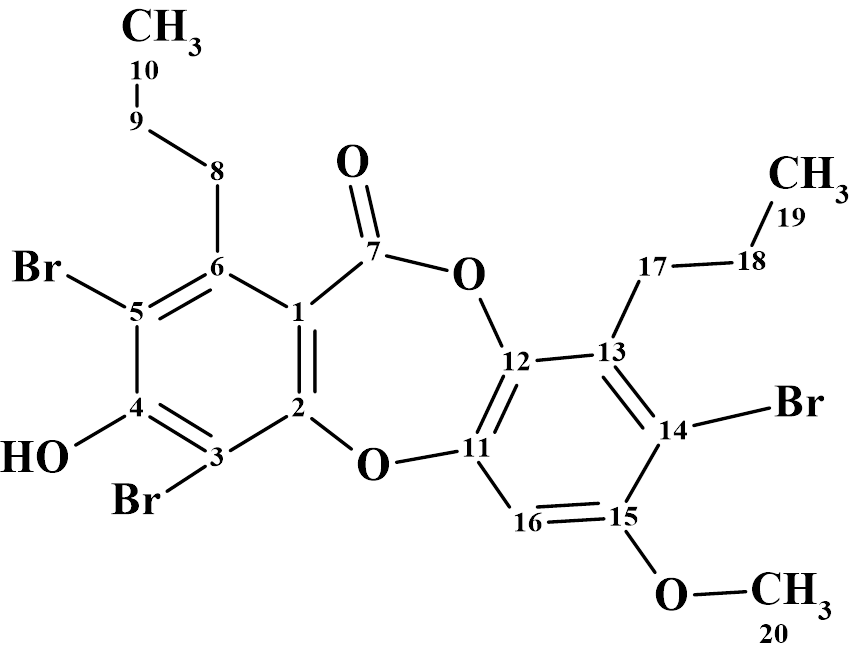
1
As we can see, this compound is characterized by a deficiency of hydrogen atoms. The molecule contains a large “silent” fragment (highlighted in red in the structure shown below), to which only one hydrogen atom is attached.
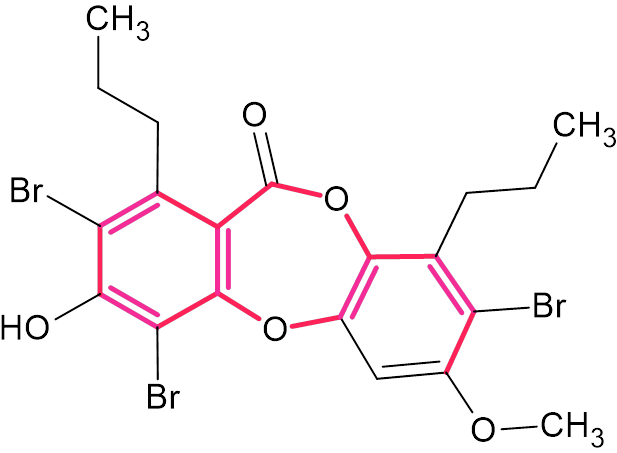
According to Crew’s rule, if the ratio of the number of hydrogen atoms to the number of skeletal atoms in a molecule is less than one, then significant difficulties typically arise in elucidating the structure using one- and two-dimensional NMR spectroscopy. For compound 1, this ratio is 0.68, but since the two side chains are very easily identified, it is actually much smaller. Therefore, it was interesting to see how ACD/Structure Elucidator handles this problem in an “ab initio” mode, that is, under the assumption that the NMR spectra of similar compounds are not available.
Spiromastixone Z4 was isolated as white powder. The molecular formula was established from HR-ESI-MS as C20H19Br3O5. The 1D and 2D NMR spectra of compound 1 (Table 1) available from [1] were entered into the program.
Table 1. NMR spectroscopic data of spiromastixone Z4 [1]
| Label | δC | δC calc HOSE | СHn | δH | M(1H) | COSY | H to C HMBC |
| C 1 | 112.6 | 118.44 | C | ||||
| C 2 | 157.6 | 157.71 | C | ||||
| C 3 | 102.5 | 98.66 | C | ||||
| C 4 | 157.6 | 158.03 | C | ||||
| C 5 | 114.1 | 109.51 | C | ||||
| C 6 | 144.6 | 146.67 | C | ||||
| C 7 | 161.9 | 160.95 | C | ||||
| C 8 | 36.1 | 38.04 | CH2 | 2.79 | t | 1.53 | C 10, C 9, C 1, C 5, C 6 |
| C 9 | 23 | 25.55 | CH2 | 1.53 | u | 0.90, 2.79 | C 10, C 8, C 6 |
| C 10 | 14.1 | 14.28 | CH3 | 0.9 | u | 1.50, 1.53 | C 18, C 9, C 17, C 8 |
| C 11 | 150.1 | 150.33 | C | ||||
| C 12 | 136.5 | 134.82 | C | ||||
| C 13 | 135.2 | 136.59 | C | ||||
| C 14 | 110.8 | 108.18 | C | ||||
| C 15 | 153.9 | 152.95 | C | ||||
| C 16 | 103.6 | 105.42 | CH | 7.07 | S | C 14, C 12, C 11, C 15 | |
| C 17 | 32.3 | 32.62 | CH2 | 2.81 | t | 1.5 | C 19, C 18, C 14, C 13, C 12 |
| C 18 | 22.1 | 22.81 | CH2 | 1.5 | u | 0.90, 2.81 | C 19, C 17, C 13 |
| C 19 | 14.4 | 14.1 | CH3 | 0.9 | u | ||
| C 20 | 57.3 | 56.67 | CH3 | 3.85 | u | C 15 |
A Molecular Connectivity Diagram, MCD, (Figure 1) was created automatically by the program.
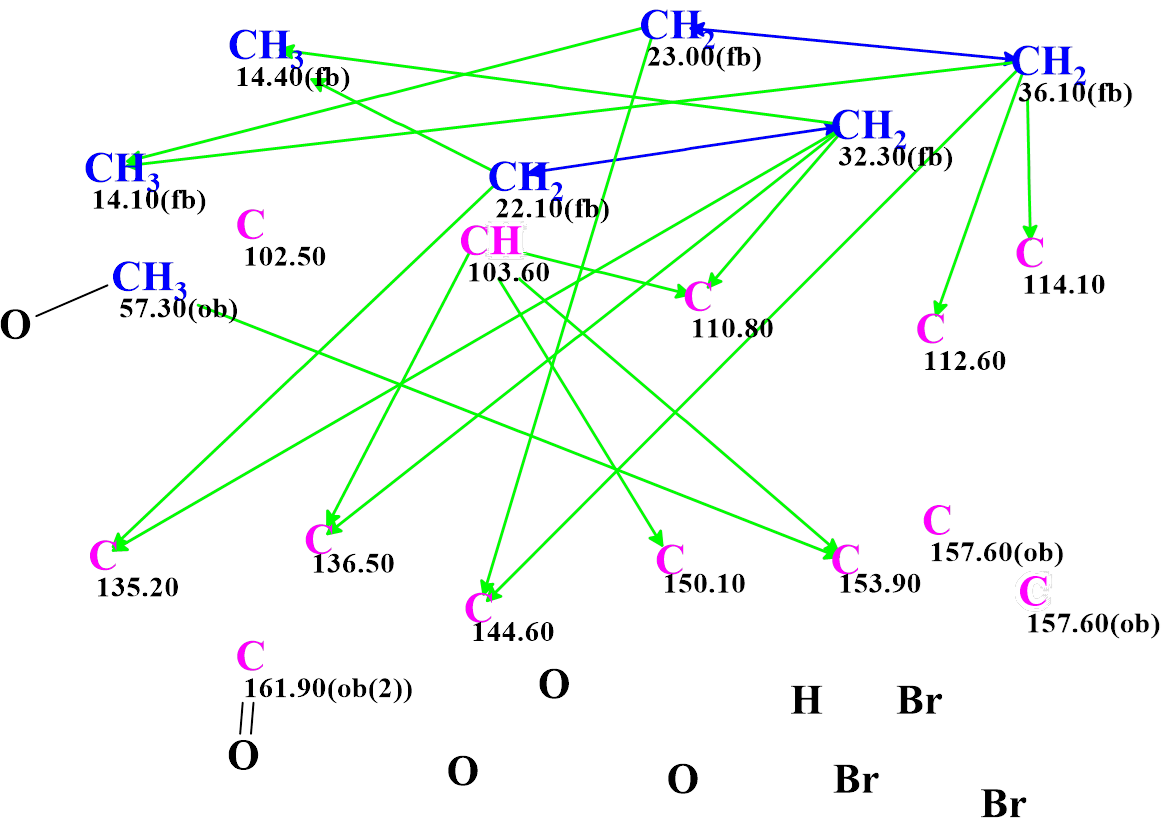
Figure 1. Molecular connectivity diagram (MCD) of spiromastixone Z4 . The hybridizations of carbon atoms are marked by corresponding colors: sp2 – violet, sp3 – blue. Labels “ob” and “fb” are set by the program to carbon atoms for which neighboring with heteroatom is either obligatory (ob) or forbidden (fb). The HMBC connectivities are marked by green arrows, while the COSY connectivities by blue arrows. The C=O bond was drawn manually to accelerate structure generation.
The IR spectrum of compound 1 (Figure 2) exhibits a strong absorption band at 1690 cm-1 which is characteristic to the stretching vibrations of the carbonyl group. Therefore, a double bond was manually drawn from the carbon atom C161.9 to the oxygen atom. The chemical shift value suggests that the carbonyl is an ester. Based on this, the C161.9 atom was supplied with the ob(2) label which indicates that two heteroatoms (in this case, two oxygen atoms) should be attached to the C161.9 atom. Given the presence of five oxygen atoms in the molecule, the “ob” label was assigned to the two C157.6 atoms. Note that both these atoms have no correlations in the HMBC, which hinders structure elucidation.
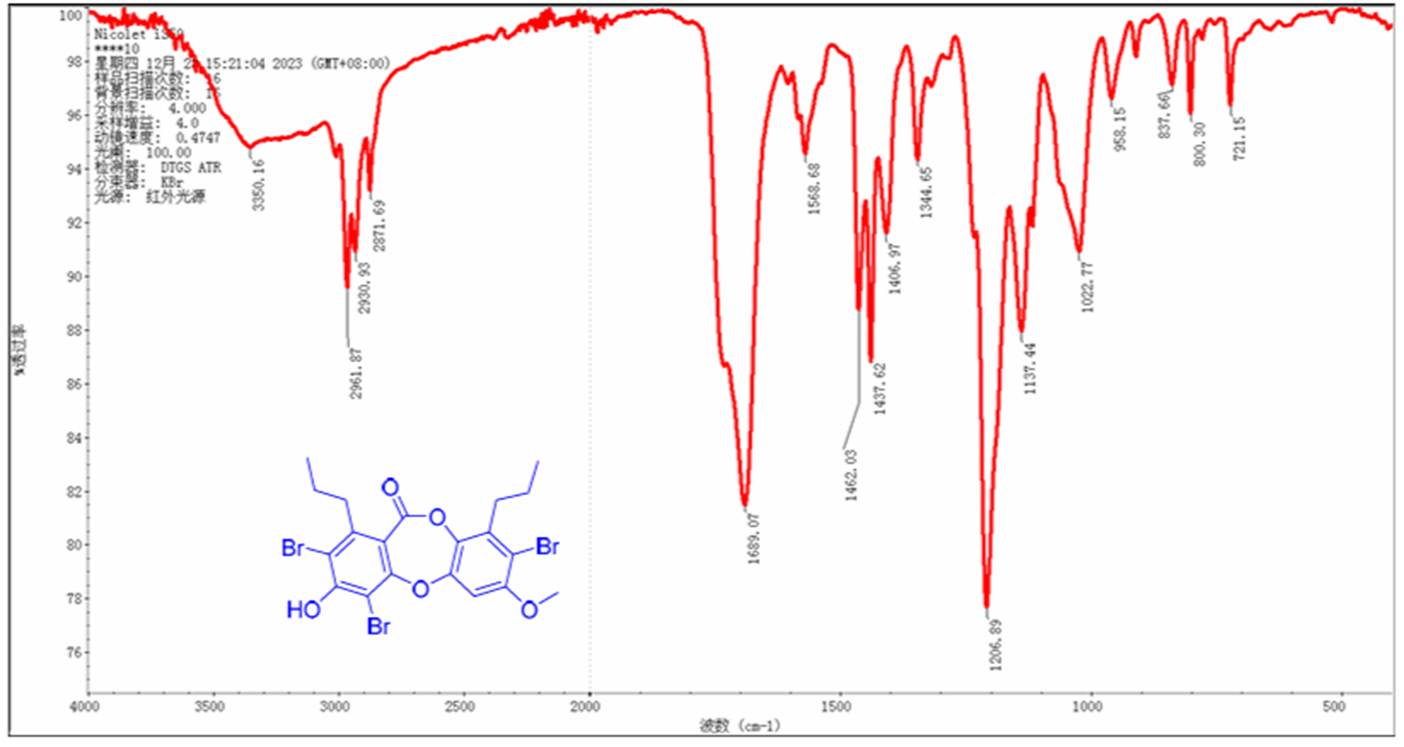
Figure 2. IR spectrum of spiromastixone Z4.
Structure generation accompanied with 13C chemical shift prediction using the empirical methods implemented into Structure Elucidator was initiated from the MCD. The generation was completed with the following results: 11,320 → (Structural Filtering) → 559 (Duplicate Removal) → 290, tg=2 min.
The output structural file was ranked in increasing order of the average deviations dA of experimental 13C chemical shifts from the ones predicted by the HOSE-code based approach. The six top ranked structures are presented in Figure 3.
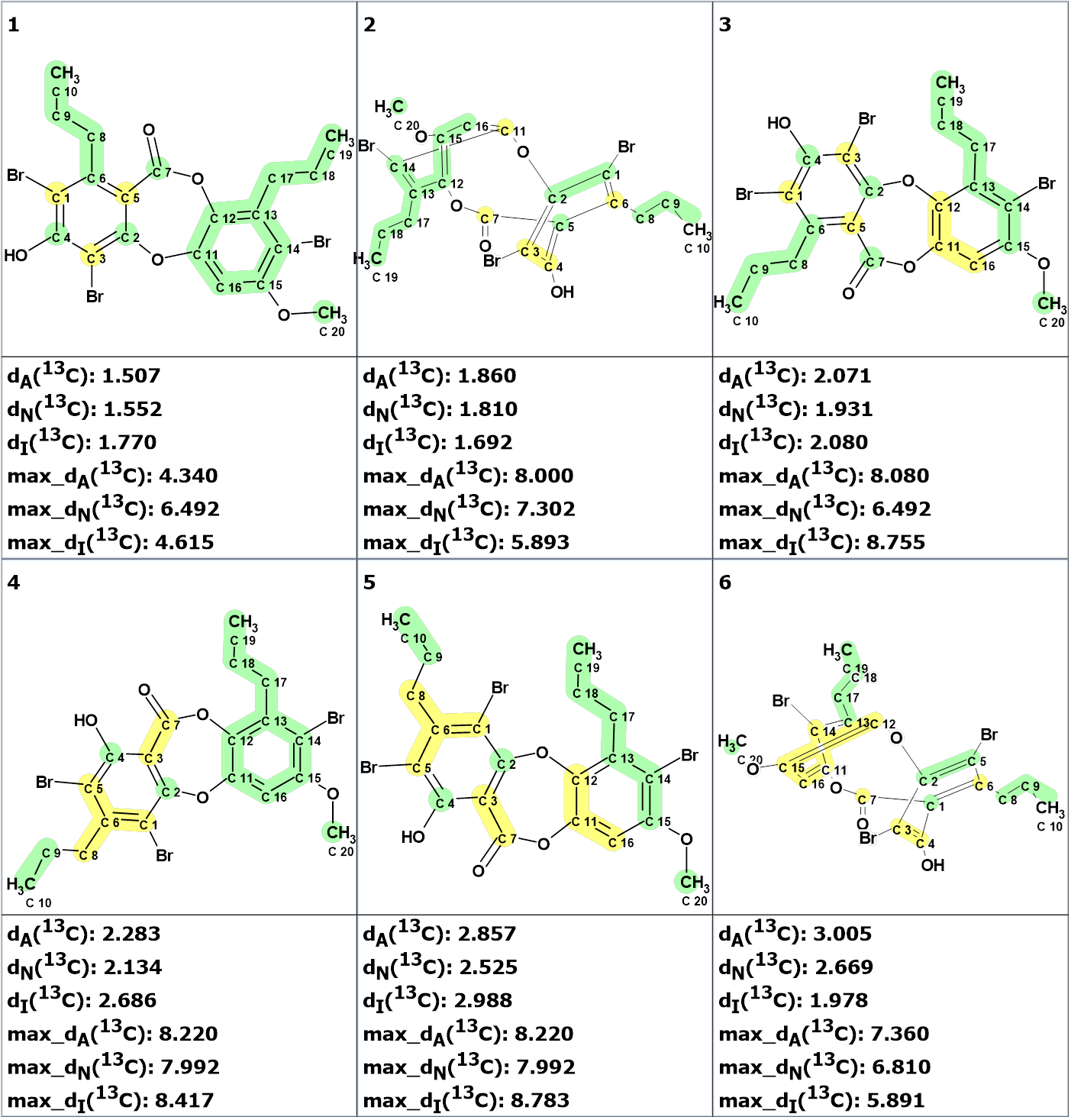
Figure 3. The six top structures of the output file. 13C chemical shift prediction was carried out using the HOSE code-based method, the neural networks, and the incremental approach. Average deviations of 13C chemical shifts determined by these methods are denoted as dA, dN and dI correspondingly. Each atom is colored to mark a difference between its experimental and calculated 13C chemical shifts. The green color represents a difference between 0 to 3 ppm, yellow was between 3 and 15 ppm.
We see that the best structure is identical to compound 1, while the atoms C1 and C5 are permuted. As shown in Figure 4, all three methods of 13C chemical shift prediction confirm the validity of the chemical shift assignment suggested by the program.
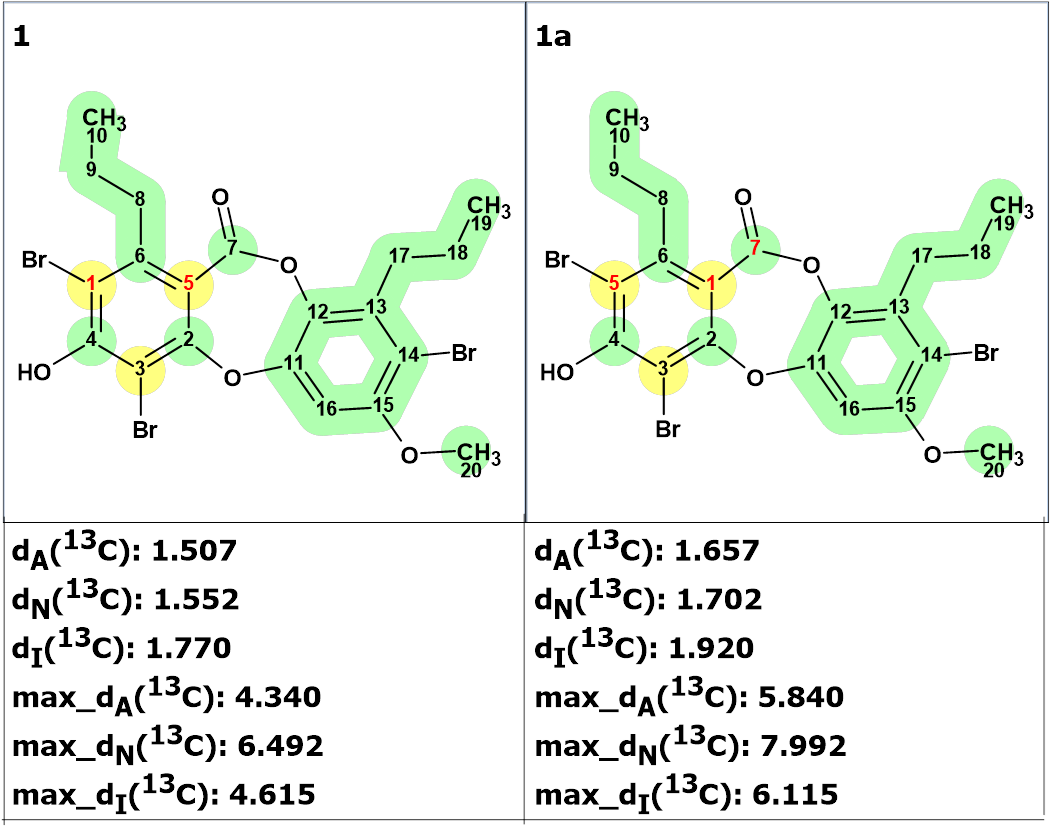
Figure 4. The 13C chemical shift assignments suggested by the program (structure 1) and by Lin et. Al. (structure 1a).
As structures 2 and 6 in Figure 3 look rather improbable chemically, the DP4 probabilities for structures 1 and 3-5 were calculated. Figure 5 shows that the best structure suggested by Structure Elucidator is confirmed correct by DP4 as well.
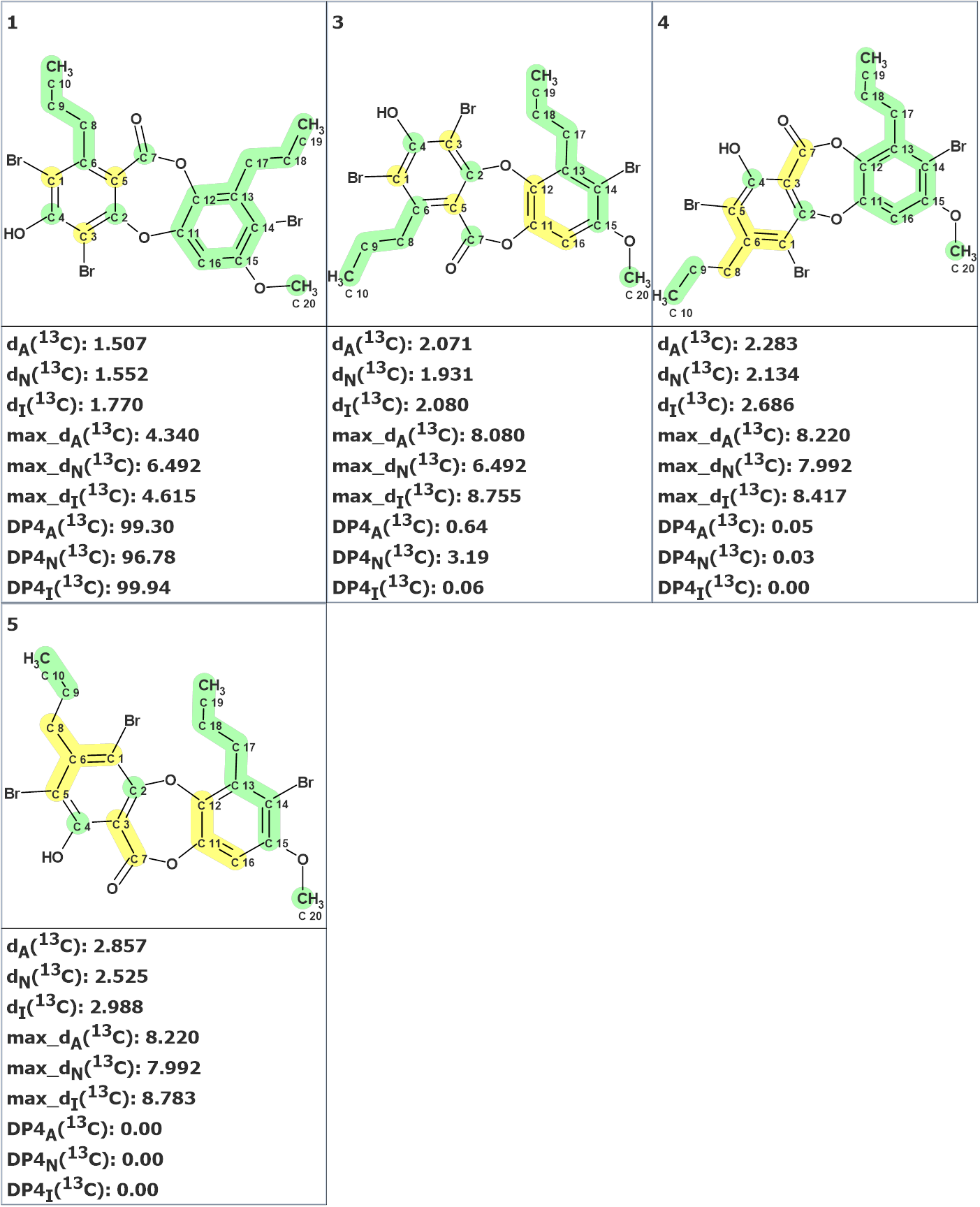
Figure 5. Probabilities DP4A, DP4N and DP4I of structure correctness calculated by the program for structures 1, 3-5.
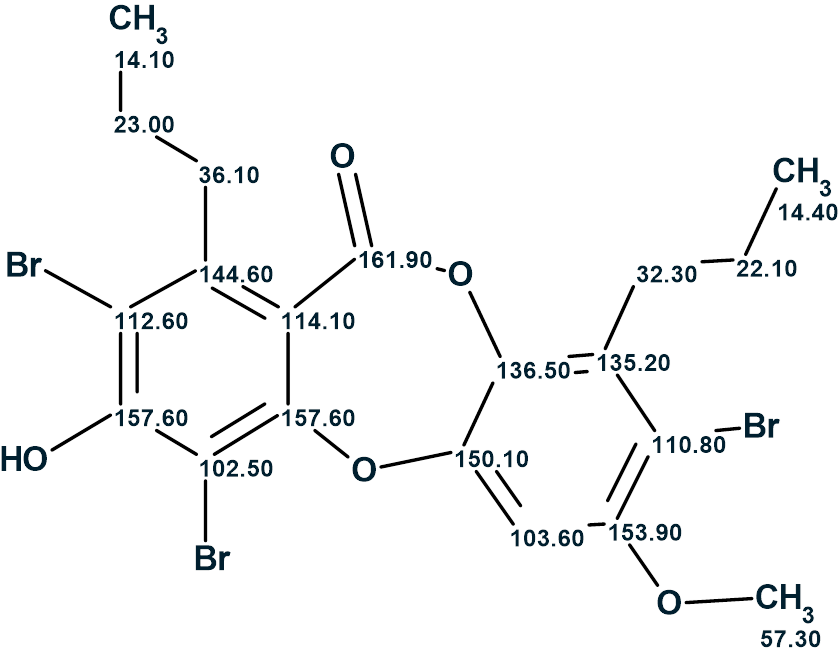
Thus, the structure of compound 1 characterized with a deficiency of hydrogen atoms was reliably determined in 2 minutes using Structure Elucidator. In so doing, the assignment of 13C chemical shifts performed by the authors [1] was corrected. The structure of Spiromastixone Z4 with the correct 13C chemical shift assignment is shown below:
References
- Huang, Z.; Liu, D.; Chen, S.;Ren, J.; Gao, C.; Li, Z.; Fan, A.; Lin, W. (2024). Brominated Depsidones with Antibacterial Effects from a Deep-Sea-Derived Fungus Spiromastix sp. Drugs, 22, 78. https://doi.org/10.3390/md22020078
