August 15, 2025
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Computer-Assisted Elucidation of Herqupenoid A Using Structure Elucidator Suite
Penicillium species are recognized for producing a wide array of structurally diverse metabolites, such as polyketides, meroterpenoids, and alkaloids. In a systematic study, Jin et al. [1] examined the secondary metabolites of a soil-derived P. herquei strain obtained from the Dianchi Lake, China, resulting in the discovery and isolation of a novel natural compound, herqupenoid A (1).
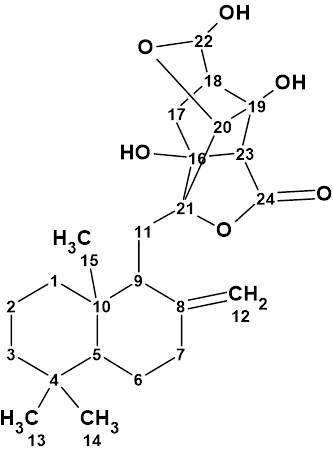
1
Compound 1 was found to possess a multicyclic, caged 2,7-dioxatetracyclo[5.4.0.04,11.05,9]hendecane fragment with an unprecedented 5/5/6/5-fused ring skeleton. Structure 1 was determined using 1D and 2D NMR and confirmed by single-crystal X-ray diffraction. Instigated by the unusual type of structure 1, we utilized the spectroscopic data published in [1] for challenging ACD/Structure Elucidator. Unfortunately, only the key HMBC and COSY correlations were shown graphically on structure 1 in the article. These data are presented in Table 1.
Table 1. NMR spectroscopic data of herqupenoid A. The overlapping 1H signals are highlighted.
| Label | δC | δC calc (HOSE) | CHn | δH | M(1H) | COSY | H to C HMBC |
| C 1 | 40 | 37.82 | CH2 | 1.77 | u | ||
| C 1 | 40 | 37.82 | CH2 | 1.21 | u | 1.77 | |
| C 2 | 20.4 | 19.17 | CH2 | 1.61 | u | 1.40, 1.77 | C 4 |
| C 2 | 20.4 | 19.17 | CH2 | 1.51 | u | ||
| C 3 | 43.4 | 42.22 | CH2 | 1.4 | u | 1.61 | |
| C 3 | 43.4 | 42.22 | CH2 | 1.21 | u | ||
| C 4 | 34.6 | 33.72 | C | ||||
| C 5 | 57 | 55.75 | CH | 1.21 | u | ||
| C 6 | 25.7 | 24.04 | CH2 | 1.33 | u | ||
| C 6 | 25.7 | 24.04 | CH2 | 1.77 | u | 1.21, 1.61, 2.39 | C 10, C 8 |
| C 7 | 39.4 | 37.84 | CH2 | 1.99 | u | ||
| C 7 | 39.4 | 37.84 | CH2 | 2.39 | u | 1.77 | |
| C 8 | 149.9 | 149.83 | C | ||||
| C 9 | 52.1 | 47.87 | CH | 2.17 | u | 1.88 | |
| C 10 | 41.1 | 39.75 | C | ||||
| C 11 | 17.5 | 31.3 | CH2 | 1.88 | u | 2.17 | C 10, C 9, C 16, C 21, C 8 |
| C 11 | 17.5 | 31.3 | CH2 | 1.99 | u | ||
| C 12 | 108.1 | 107.48 | CH2 | 4.91 | u | C 7, C 9, C 8 | |
| C 12 | 108.1 | 107.48 | CH2 | 4.83 | u | ||
| C 13 | 22.1 | 22.4 | CH3 | 0.83 | s | C 14, C 4, C 3 | |
| C 14 | 34.1 | 34.26 | CH3 | 0.89 | s | C 4, C 5 | |
| C 15 | 15 | 22.45 | CH3 | 0.72 | s | C 1, C 9, C 5 | |
| C 16 | 87 | 85.63 | C | ||||
| C 17 | 33.5 | 36.9 | CH2 | 1.51 | u | ||
| C 17 | 33.5 | 36.9 | CH2 | 2.06 | u | 2.55 | C 23, C 16, C 21 |
| C 18 | 51.3 | 52.05 | CH | 2.55 | u | 2.06, 5.30 | C 23 |
| C 19 | 88.7 | 91.22 | C | ||||
| C 20 | 86.9 | 83.37 | CH | 3.81 | u | C 18, C 23, C 16, C 19, C 21 | |
| C 21 | 93.9 | 88.32 | C | ||||
| C 22 | 105.9 | 105.98 | CH | 5.3 | u | 2.55 | C 17, C 20, C 19 |
| C 23 | 61.3 | 58.83 | CH | 2.88 | u | C 21, C 24 | |
| C 24 | 176.1 | 174.23 | C |
We see that many overlapping signals are observed in the 1H spectrum, which makes the problem even more difficult. Compound 1 was obtained as a colorless crystal. Its molecular formula, C24H34O6, was determined by the positive HR-ESI-MS data (m/z 441.2273 [M + Na]+, calcd. for C24H34O6Na+, 441.2248), which suggested 8 indexes of hydrogen deficiency.
The molecular formula and available spectroscopic data (Table 1) were entered into the program which created the molecular connectivity diagram, MCD shown in Figure 1.

Figure 1. Molecular connectivity diagram (MCD) of herqupenoid A. The hybridizations of carbon atoms are marked by the corresponding colors: sp2 – violet, sp3 – blue, not sp – light blue. The labels “ob” and “fb” are set by the program to carbon atoms for which neighboring with a heteroatom is either obligatory (ob) or forbidden (fb). HMBC connectivities are marked by green arrows, while COSY connectivities by blue arrows. Ambiguous connectivities are marked by a dotted line.
We see that the presence of overlapping signals in the 1H spectrum led to producing ambiguous COSY correlations. In addition, there are four carbon atoms (C 87, C 88.7, 93.9, and 105.9) whose hybridizations may be either sp3 or sp2. Therefore, both possible hybridizations must be tried during structure generation. No user edits of MCD were done.
Checking the MCD for the presence of contradictions in the HMBC and COSY data showed that the data are consistent, so strict structure generation was initiated which was completed with the following results: k = 723 → (Structure Filtering) → 56 → (Duplicate Removal) → 30, tg=7s. 13C chemical shifts were predicted for all structures of the output file and the structures were ranked in increasing order of average deviations of the predicted chemical shifts from experimental ones. The nine top ranked structures are presented in Figure 2.
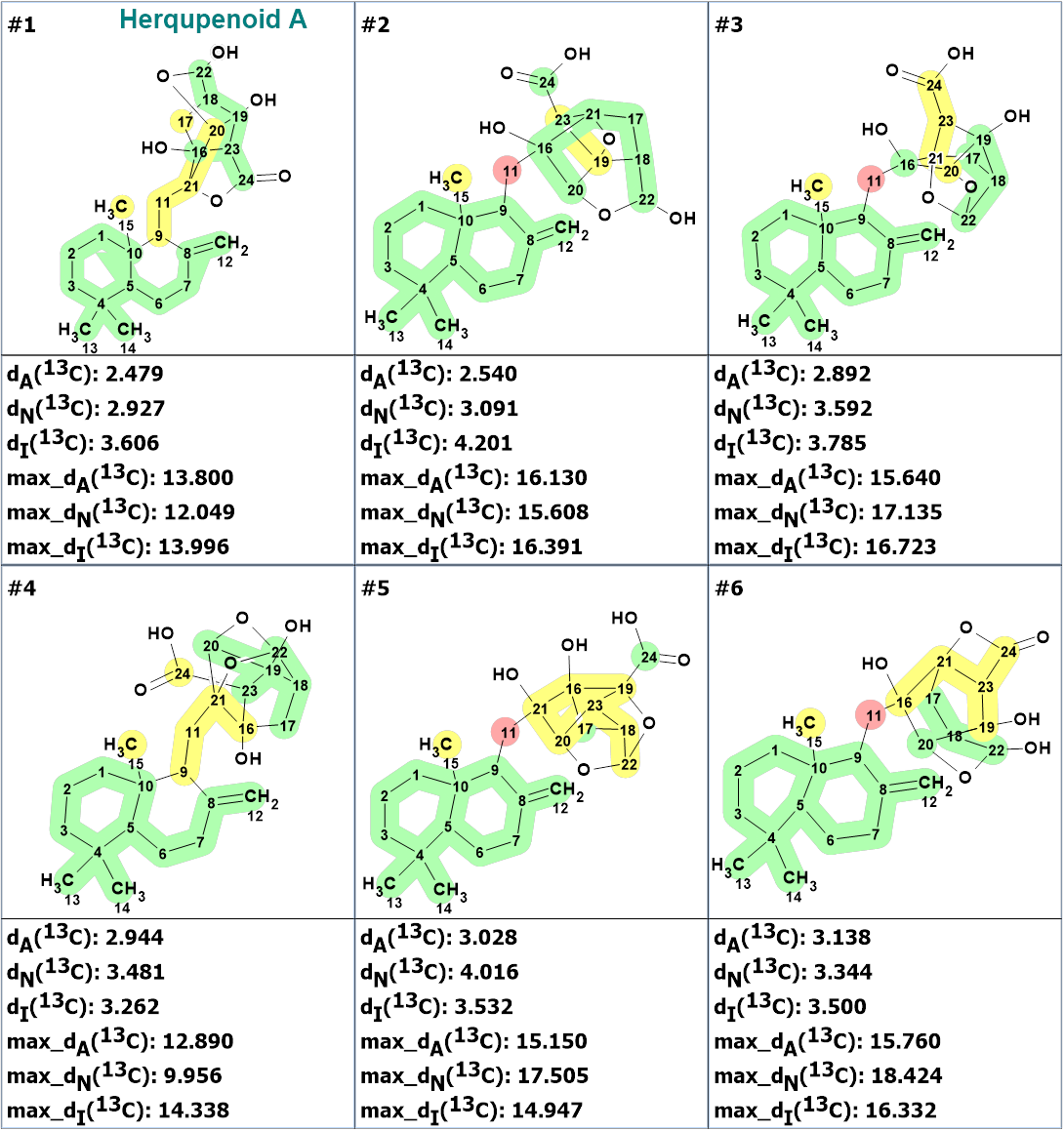

Figure 2. The nine top ranked structures of the output file. 13C chemical shift prediction was carried out using the HOSE code-based method, the neural networks, and the incremental approach. Average deviations of 13C chemical shifts determined by these methods are denoted as dA, dN and dI correspondingly. Each atom is colored to mark a difference between its experimental and calculated 13C chemical shifts. The green color represents a difference between 0 to 3 ppm, yellow was between 3 and 15 ppm and red more than 15 ppm. The structures are ranked in increasing order of dA values.
The figure shows that the first ranked structure #1 is herqupenoid A. The other structures should be rejected both from their larger values of dA and from the deviations from the IR spectrum of compound 1 (Figure 3).
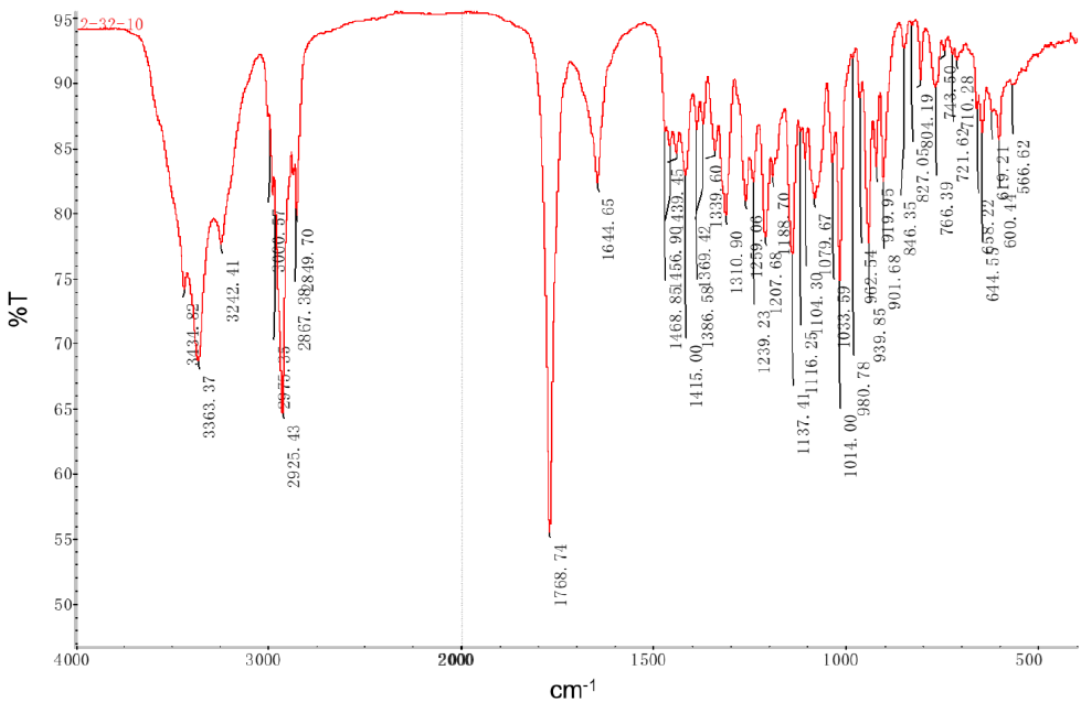
Figure 3. The Infrared spectrum of herqupenoid A.
The carbonyl band observed at 1768 cm-1 is characteristic for five-membered lactones and cannot be explained by vibrations of OH-C=O group (~1700 cm-1). Moreover, no typical features of an acid group are seen around 3000 cm-1.
Therefore, an unprecedented structure of herqupenoid A was elucidated with the help of Structure Elucidator, fully automatically in 7 seconds. Structure 1 with the 13C shifts assigned by the program is shown below:

References
- Jin, Y. Zhao, H. Wang, R. Jiang, J. Wei, H. Zeng, W Sun, Y. Zhang, Z. Hu. (2024). Herqupenoid A, an Unparalleled Sesquiterpene-Quinone Hybrid Featuring a Multicyclic Caged 2,7-Dioxatetracyclo[5.4.0.04,11.05,9]Hendecane Fragment from Penicillium herque. Org. Lett., 26, 10146−10151. DOI: 10.1021/acs.orglett.4c03867
