May 1, 2015
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Bacillusin A
Bacillus, a genus of Gram-positive bacteria, have been screened for antimicrobial activities over the past few decades. Many Bacillus spp. have been shown to produce antibiotic polyketides such as macrolactins, difficidins, and oxidifficidins, as well as lipopeptides such as surfactins, iturins, and fengycins. Some Bacillus spp. have also been used as biocontrol agents against plant and animal pathogens in agriculture and aquaculture. R.R. Ravu and coworkers [1] performed work searching for new antibiotics against drug-resistant bacteria. The B. amyloliquefaciens subsp. plantarum strain AP183 was shown to produce potent activity against methicillin-resistant Staphylococcus aureus (MRSA) (IC50 of <10 µg/mL). Preliminary fractionation of the crude extracts afforded activity-enriched fractions that contained an apparently unknown active compound with strong UV absorptions. Thus, a scale-up fermentation of this Bacillus strain was conducted, leading to the isolation and identification of a new antibacterial macrocyclic polyene antibiotic compound (1), designated Bacillusin A.
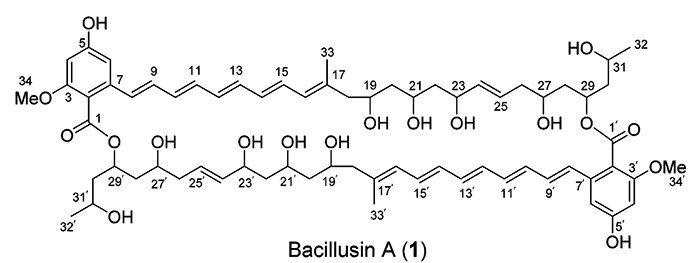
Compound 1 was obtained as a yellow powder and exhibited a molecular ion peak at m/z 1219.6172 for [M + Na]+ in the high-resolution ESIMS spectrum. The 13C NMR, DEPT, and HSQC spectra in DMSO-d6 showed 34 carbon resonances, including two methyl groups, one methoxy group, six methylene groups, six oxymethine groups, 13 olefinic or aromatic tertiary carbons, five olefinic or aromatic quaternary carbons, and one carboxylic ester carbon. The resonance at dC 131.5 correlating with two olefinic protons at dH 6.80 and 6.32 in the HSQC spectrum represented two carbons. Thus, compound 1 should be a symmetric dimer with a molecular formula of C68H92O18 containing 86 skeleton atoms.
The NMR data used for structure elucidation of compound 1 with the aid of ACD/Structure Elucidator Suite are presented in Table 1.
Table 1: NMR spectroscopic data [1] for Bacillusin A.
| Label | δC | δC calc | XHn | δH | M(J) | C HMBC |
| C 1 | 167.3 | 167.73 | C | |||
| C 2 | 115 | 114.95 | C | |||
| C 3 | 157.3 | 157.97 | C | |||
| C 4 | 98.7 | 97.96 | CH | 6.34 | s | C 5, C 1, C 6, C 3, C 2 |
| C 5 | 159.3 | 162.18 | C | |||
| C 6 | 102.5 | 109.01 | CH | 6.63 | s | C 2, C 1, C 5, C 8, C 4 |
| C 7 | 136.1 | 141.91 | C | |||
| C 8 | 127.9 | 129.04 | CH | 6.38 | d | C 9, C 7, C 2, C 6 |
| C 9 | 131.5 | 130.1 | CH | 6.8 | dd | C 7, C 11 |
| C 10 | 135.7 | 130.78 | CH | 6.48 | dd | C 8, C 11 |
| C 11 | 132.1 | 131.44 | CH | 6.35 | dd | C 10 |
| C 12 | 131.5 | 129.83 | CH | 6.32 | dd | C 10, C 14, C 11 |
| C 13 | 135.1 | 128.14 | CH | 6.36 | dd | C 11, C 12 |
| C 14 | 130.5 | 129.71 | CH | 6.16 | dd | C 15, C 16 |
| C 15 | 130.7 | 133.26 | CH | 6.46 | dd | C 14, C 17 |
| C 16 | 126.9 | 127.69 | CH | 5.82 | d | C 18, C 15, C 33 |
| C 17 | 138.6 | 136.78 | C | |||
| C 18 | 48.1 | 46.91 | CH2 | 2 | u | C 16, C 33, C 20, C 17, C 19 |
| C 18 | 48.1 | 46.91 | CH2 | 2.08 | u | |
| C 19 | 67.7 | 67.84 | CH | 3.74 | u | |
| C 20 | 44.6 | 46.24 | CH2 | 1.37 | u | C 22, C 18, C 21 |
| C 21 | 67.6 | 65.09 | CH | 3.67 | u | C 3 |
| C 22 | 45.1 | 45.38 | CH2 | 1.5 | u | |
| C 22 | 45.1 | 45.38 | CH2 | 1.43 | u | C 24, C 20, C 23, C 21 |
| C 23 | 69.7 | 67.24 | CH | 4.03 | u | C 25, C 22, C 21 |
| C 24 | 135.7 | 136.51 | CH | 5.37 | dd | C 26, C 25, C 1, C 23 |
| C 25 | 126.3 | 128.43 | CH | 5.51 | dt | C 23, C 24, C 26, C 27 |
| C 26 | 41.1 | 37.15 | CH2 | 1.98 | u | C 28, C 27, C 24, C 25 |
| C 26 | 41.1 | 37.15 | CH2 | 2.04 | u | |
| C 27 | 66.3 | 67.17 | CH | 3.56 | u | |
| C 28 | 42.2 | 43.44 | CH2 | 1.63 | u | C 29 |
| C 29 | 70.2 | 72.55 | CH | 5.37 | u | C 1 |
| C 30 | 44.7 | 45.08 | CH2 | 1.58 | u | C 28 |
| C 31 | 62.5 | 64.35 | CH | 3.74 | u | |
| C 32 | 24.3 | 24.17 | CH3 | 1.05 | d | C 31, C 30 |
| C 33 | 17.2 | 24.64 | CH3 | 1.69 | s | C 15, C 16, C 18, C 17 |
| C 34 | 55.5 | 55.49 | CH3 | 3.67 | s | C 3 |
We see that overlapping chemical shifts present both in 13C and 1H NMR spectra (pairs of chemical shift are indicated by color in Table 1). Therefore we can expect that ambiguous connectivities will exist in the data set, which can hamper structure elucidation.
A Molecular Connectivity Diagram (MCD) created from the spectroscopic data in Table 1 is presented in Figure 1. As the number of HMBC correlations is high, Figure 1 does not allow for detailed visual analysis of the atom properties and connectivities. Thus in Figure 2, the atoms are presented with only their chemical shifts and properties.
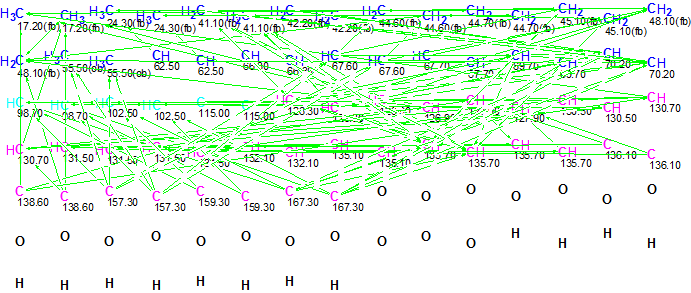
Figure 1. Initial Molecular Connectivity Diagram for Bacillusin A. All connectivities are shown.
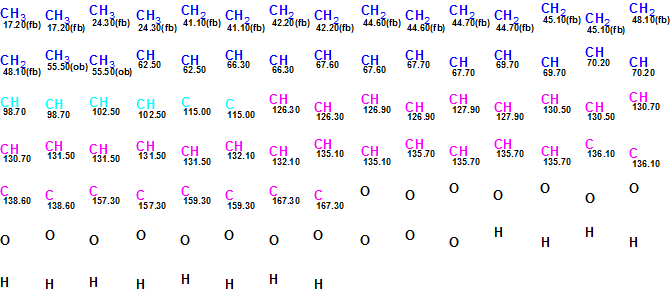
Figure 2. Initial Molecular Connectivity Diagram for Bacillusin A. Only atoms and their 13C chemical shifts are shown.
MCD overview. The initial MCD contains six light blue carbon atoms (their hybridization is not sp) whose 13C chemical shifts are observed in the region 98.70 – 115.00 ppm. Following the authors’ conclusions, hybridization of these atoms was set as sp2). The 13C chemical shifts of the carbon atoms CH 62.50 – C 70.20 and the1H chemical shifts of the attached hydrogens allow for us to recognize these atoms as oxygenated, and thus supply them with the label “ob” (at least one heteroatom is a neighbor). The same label was also assigned to the sp2 hybridized carbons C 157.3 – C 167.3. The numbers of hydrogens attached to neighbor carbons were set in accordance with 1H multiplicities shown in Table 1. The resulting edited MCD is presented in Figure 3.
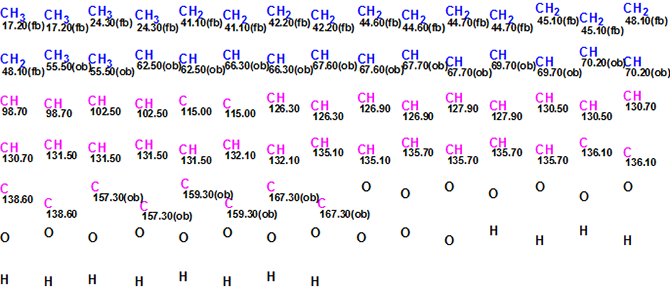
Figure 3. Edited Molecular Connectivity Diagram for Bacillusin A. Only atoms and their 13C chemical shifts are shown.
After checking the MCD, the program produced indicated that at least one non-standard connectivity was present. Inspection of the HMBC spectrum from the original Supporting Information [1] (see an HMBC fragment, Figure 4) demonstrated that the intensities of peaks corresponding to correlations 6.34 to 167.20 and 6.63 to 167.20 are quite small, suggesting that that these correlations are of nonstandard length.
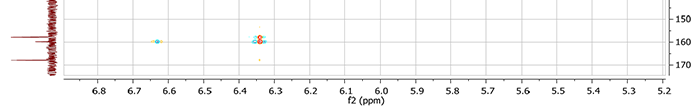
Figure 4. Fragment of the HMBC spectrum for Bacillusin A. It is evident that intensities of correlations 6.34 to 167.20 and 6.63 to 167.20 are quite small.
As the total number of HMBC correlation is high, it is rational to remove these “suspicious” connectivities in this case (otherwise their lengths can be set as 2-4). After removing these connectivities, MCD checking detected again the presence of nonstandard connectivities. Consequently Fuzzy Structure Generation was initiated in the mode “Determine Options Automatically”. The results were: k = 2492 → 242 → 242, tg = 15 s, 1 (from 53) correlation having been extended during generation, with 4 (from 53 possible) connectivity combinations used.
The first four structures of the output file, ranked in increasing order of an average deviation dA(13C), are presented in Figure 5.
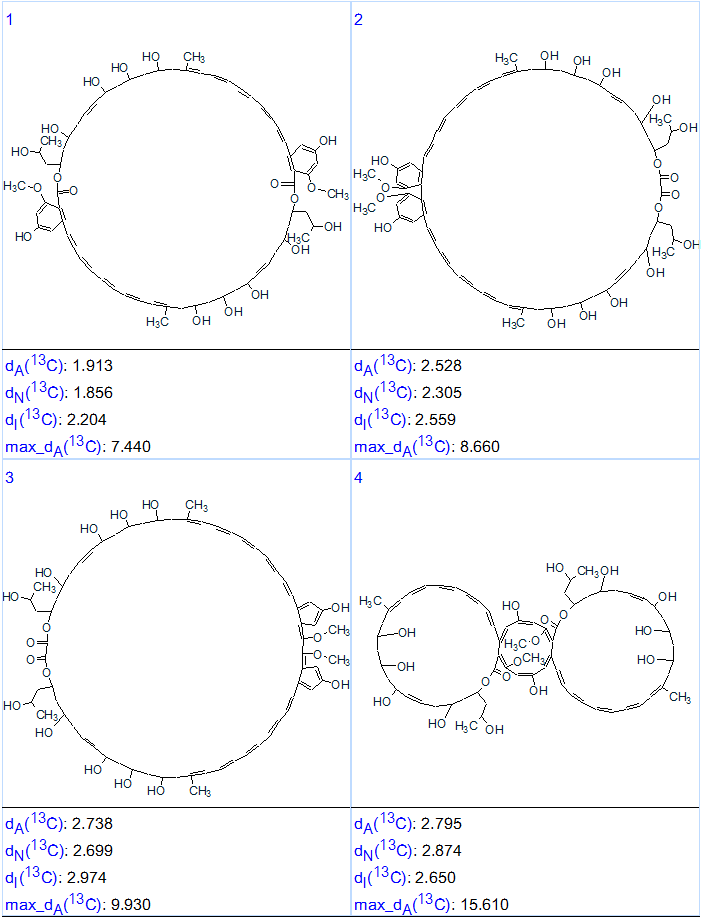
Figure 5. The first four structures of the ranked output file for Bacillusin A.
By comparing structure #1 of the ranked file with structure 1 we can conclude that both structures are identical. The elucidated structure of Bacillusin A (2) is shown below.
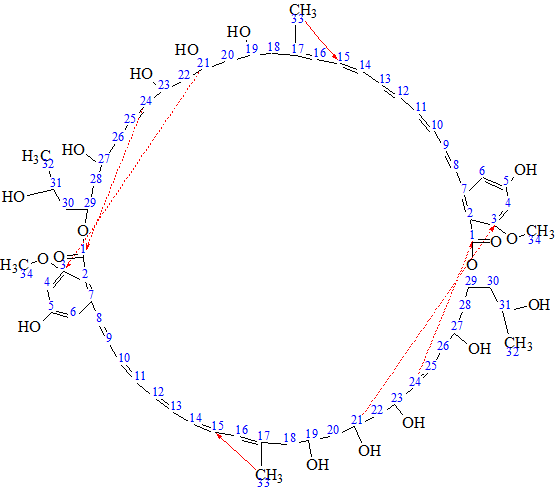
2
Solid red arrows show nonstandard connectivities 1.69 to 130.7 (see structure 1/b> and Table 1), while ambiguous connectivities are marked by dotted lines. It is interesting to note that correlation 1.69 → 130.7 (H 33 → C 15) could not be related to the “suspicious” one from its intensity. This is seen in Figure 6 where corresponding fragment of HMBC spectrum is presented.
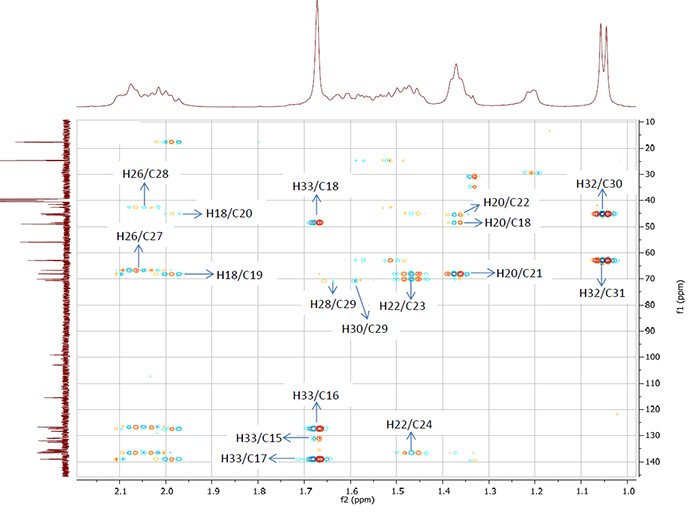
Figure 6. Fragment of the HMBC spectrum for Bacillusin A. The intensity of the correlation 1.69→130.7 (H 33 / C 15) is not less than those of other correlations, all of which are of standard lengths.
It is worthy to remind that, in general case, structure generation from 2D NMR data becomes very challenging when a molecule under investigation possesses elements of symmetry [2]. This difficulty was overcome by using Structure Elucidator Suite with only minor manual modifications needed. As we see, Fuzzy Structure Generation was able to elucidate the structure of a large molecule containing 86 skeletal atoms (and in the presence of ambiguous connectivities) in only 15 seconds.
References
- R.R. Ravu, M.R. Jacob, X. Chen, M. Wang, S. Nasrin, J.W. Kloepper, M. R. Liles, D. A. Mead, I. A. Khan, X.-C. Li. Bacillusin A, an Antibacterial Macrodiolide from Bacillus amyloliquefaciens AP183.. J. Nat. Prod., DOI: 10.1021/np500911k, Publication Date (Web): March 10, 2015.
- M.E. Elyashberg, A.J. Williams, Blinov K.A. Contemporary Computer-Assisted Approaches to Molecular Structure Elucidation, Cambridge, RSC Publishing, 2012, 482 p. http://pubs.rsc.org/en/Content/eBook/978-1-84973-432-5


