July 22, 2025
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Computer-Assisted Elucidation of Canumycin C Using Structure Elucidator Suite
Napyradiomycins are meroterpenoid natural products characterized by a naphthoquinone core decorated with prenyl and geranyl side chains. These hybrid molecules have garnered considerable interest due to their potent antibacterial properties. Since their initial discovery in Streptomyces rubra in 1986, more than 50 structurally diverse analogs have been isolated from various actinomycetes. A consistent feature among them is the presence of the retained naphthoquinone scaffold. Hao et al. [1] recently reported the identification of five new macrocyclic napyradiomycins from Streptomyces canus SJ-019, including three compounds with unprecedented skeletons. One such compound, Canumycin C (1), features a dearomatized core bridged by a distinctive double-arch structure.
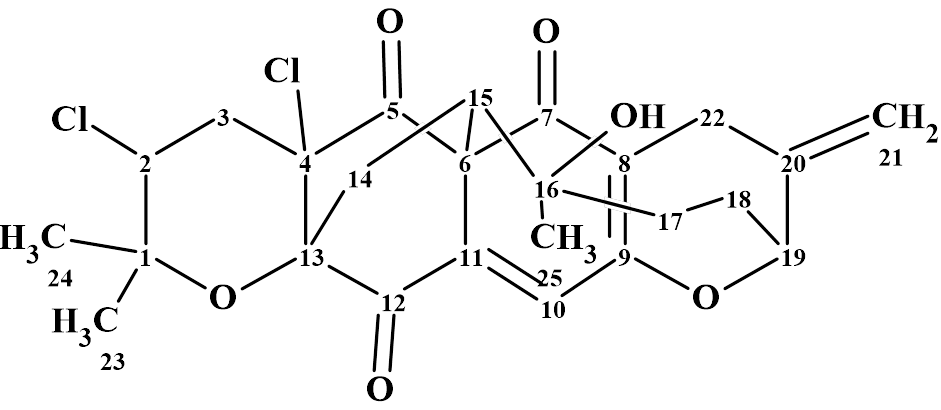
1
Canumycin C differs from other napyradiomycins in its high degree of cyclization and hence an unusual double-arch bridge topology. Another striking difference between 1 and other napyradiomycins is the dearomatization of the naphthoquinone core, as the highly conjugated naphthoquinone ring is typical in this class of metabolites.
Canumycin C was isolated as a colorless powder. HR-ESI-MS analysis gave a molecular formula of C25H26Cl2O6, indicating 12 degrees of unsaturation. The molecular formula and NMR spectroscopic data (Table 1) of Canumycin C were used by us for testing ACD/Structure Elucidator.
Table 1. 1D and 2D NMR spectroscopic data of Canumycin C [1].
| Label | δC | δCcalc (HOSE) | δCHn | δH | M(1H) | COSY | H to C HMBC |
| C 1 | 80.1 | 80.62 | C | ||||
| C 2 | 58.7 | 58.23 | CH | 4.48 | u | 2.75 | C 24, C 23, C 3, C 4, C 1 |
| C 3 | 36.7 | 41.77 | CH2 | 1.89 | u | ||
| C 3 | 36.7 | 41.77 | CH2 | 2.75 | u | 4.48 | C 2, C 4, C 1, C 13, C 5 |
| C 4 | 70.5 | 70.31 | C | ||||
| C 5 | 192.3 | 197 | C | ||||
| C 6 | 60.4 | 72.32 | C | ||||
| C 7 | 184.1 | 193.03 | C | ||||
| C 8 | 121.7 | 115.33 | C | ||||
| C 9 | 162.7 | 163.89 | C | ||||
| C 10 | 131.1 | 119.24 | CH | 6.99 | s | C 6, C 8, C 11, C 9, C 12 | |
| C 11 | 138.8 | 133.42 | C | ||||
| C 12 | 194.3 | 189.99 | C | ||||
| C 13 | 83.1 | 85.07 | C | ||||
| C 14 | 35.3 | 30.77 | CH2 | 2.73 | u | 3.93 | C 15, C 6, C 4, C 13, C 16, C 12 |
| C 14 | 35.3 | 30.77 | CH2 | 2.2 | u | ||
| C 15 | 43.8 | 43.22 | CH | 3.93 | d | 2.73 | C 25, C 14, C 17, C 6, C 16, C 11, C 5 |
| C 16 | 86.3 | 78.73 | C | ||||
| C 17 | 36.6 | 34.52 | CH2 | 1.87 | u | 1.78 | |
| C 17 | 36.6 | 34.52 | CH2 | 2.39 | u | C 18, C 15, C 19, C 16 | |
| C 18 | 27.4 | 28.35 | CH2 | 1.84 | u | ||
| C 18 | 27.4 | 28.35 | CH2 | 1.78 | u | 1.87, 4.71 | C 17, C 19, C 16, C 20 |
| C 19 | 80.5 | 81.47 | CH | 4.71 | u | 1.78 | C 22, C 18, C 17, C 21, C 20, C 9 |
| C 20 | 139 | 141.05 | C | ||||
| C 21 | 113.8 | 111.9 | CH2 | 5.18 | u | C 22, C 19, C 20 | |
| C 21 | 113.8 | 111.9 | CH2 | 5.16 | u | ||
| C 22 | 23.1 | 34.57 | CH2 | 3.14 | u | ||
| C 22 | 23.1 | 34.57 | CH2 | 3.32 | u | C 19, C 21, C 8, C 20, C 9, C 7 | |
| C 23 | 29.6 | 27.68 | CH3 | 1.53 | s | C 24, C 2, C 1 | |
| C 24 | 23.4 | 21.01 | CH3 | 1.46 | s | C 23, C 2, C 1 | |
| C 25 | 15.2 | 25.92 | CH3 | 0.71 | s | C 17, C 15, C 16 |
Although the molecular formula shows that Canumycin is a hydrogen deficient compound, there are no large “silent” fragments in the molecule and the hydrogen atoms are fairly evenly distributed throughout the structure. As a result, the number of HMBC correlations is quite large, namely 60.
The NMR spectroscopic data were entered into the program, and a Molecular Conenctivity Diagram (MCD) was created (Figure 1).

Figure 1. The molecular connectivity diagram (MCD) of Canumycin C. The hybridizations of carbon atoms are marked by corresponding colors: sp2 – violet, sp3 – blue, not sp (sp3 or sp2) – light blue. The “fb” labels are set by the program to carbon atoms for which neighboring with heteroatom is forbidden (fb). The HMBC connectivities are marked by green arrows, while COSY connectivities are marked by blue arrows.
The MCD contains six light blue carbon atoms which can be sp3 or sp2 hybridized. All the possibilities for these will be explored during structure generation. Atoms 184.1, 192.3, and 194.3 are obviously carbonyls, but no manual editing of the atomic properties was done in the MCD. Structure generation was initiated which was completed in 12 min with the following result: k= 1397 → (Structure Filtering) → 107 → (Duplicate Removal) à107.
Prediction of the 13C chemical shifts was performed for all structures of the output file using the three methods provided by the ACD/SE system: the HOSE code-based method, the neural networks, and the incremental approach. The structures were then ranked in order of increasing average deviation dA of the predicted chemical shifts from the experimental values.
The top twelve structures of the ranked output file are shown in Figure 2.
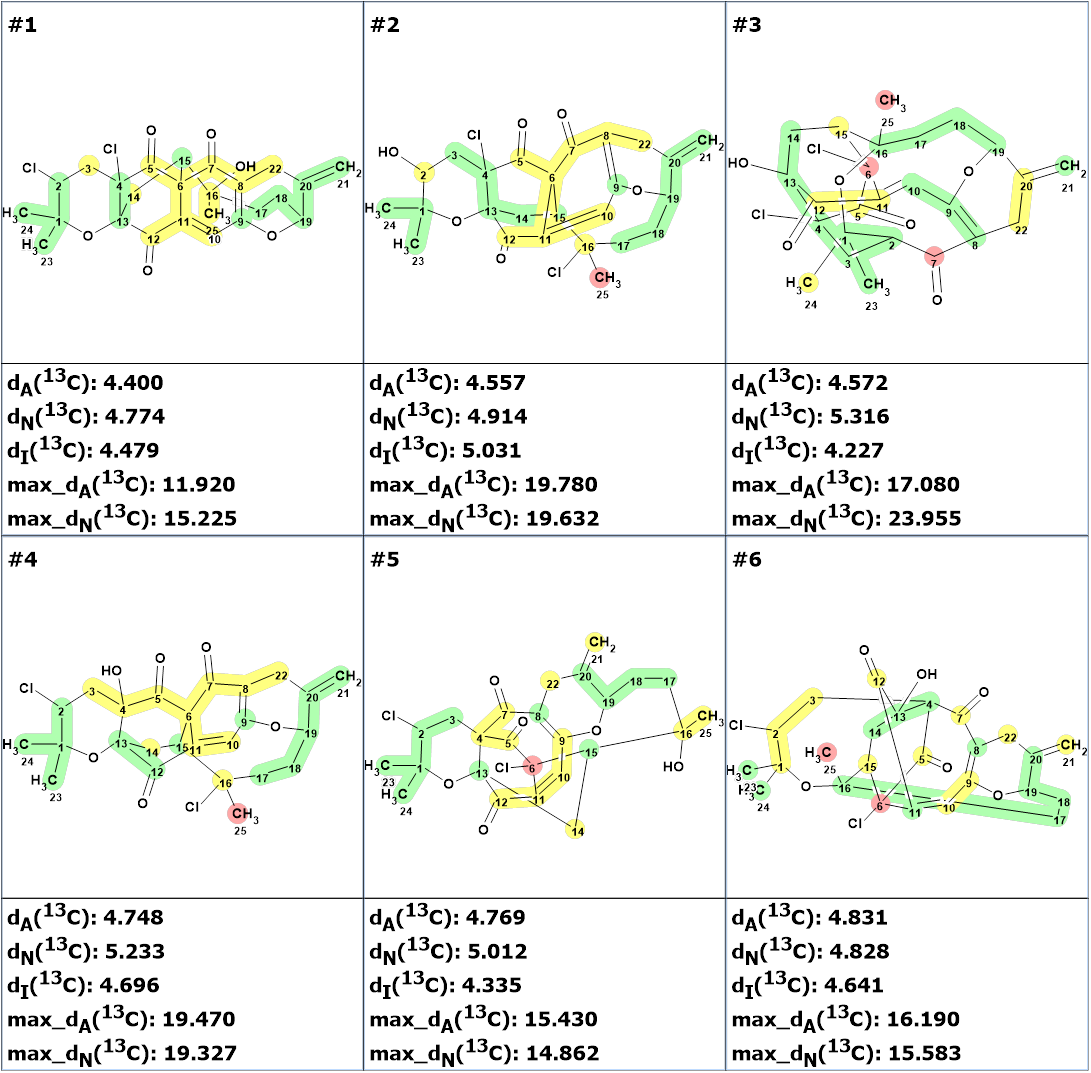
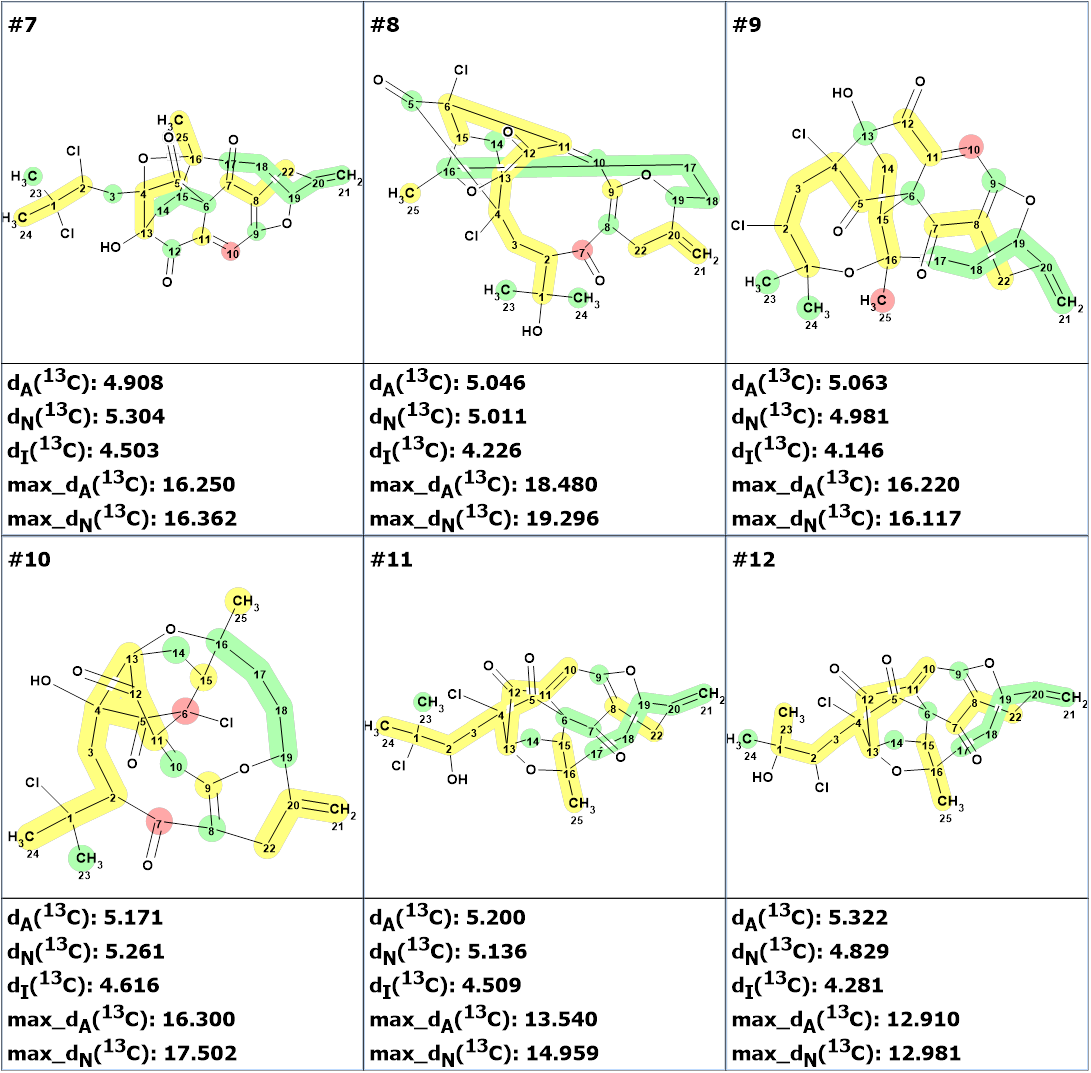
Figure 2. The ranked output file. 13C chemical shift prediction was carried out using the HOSE code-based method, the neural networks, and the incremental approach. Average deviations of 13C chemical shifts determined by these methods are denoted as dA, dN and dI correspondingly. Each atom is colored to mark a difference between its experimental and calculated 13C chemical shifts. The green color represents a difference between 0 to 3 ppm, yellow was between 3 and 15 ppm, and red more than 15 ppm.
We see that structure #1 coincides with Canumycin C structure proposed by the authors of the work [1]. However, the values of the average deviations turned out to be significantly greater than those values that are usually typical for a correct structure (1.5-2.5 ppm). Such large values can be observed in very rare cases and for unusual structures, when similar fragments cannot be found in the ACD/Labs NMR Predictors training database. In these cases, prediction errors can be significantly greater than usual. The validity of structure #1 is confirmed by the fact that the other structures have higher values of average and maximum deviations. The presence of atoms colored red shows that these structures are less probable. However, given the unusual nature of structure 1 and the large deviation values, its validity would preferably be confirmed by X-ray analysis if obtaining monocrystal is possible or by DFT calculations of the chemical shifts.
It is interesting to note that when the evident three carbonyl groups were drawn manually in the MCD, the same 107 structures were generated in 40s that is about 20 times faster. This example demonstrates the usefulness of imposing evident constraints that deliberately cannot lead to mistaken solutions.
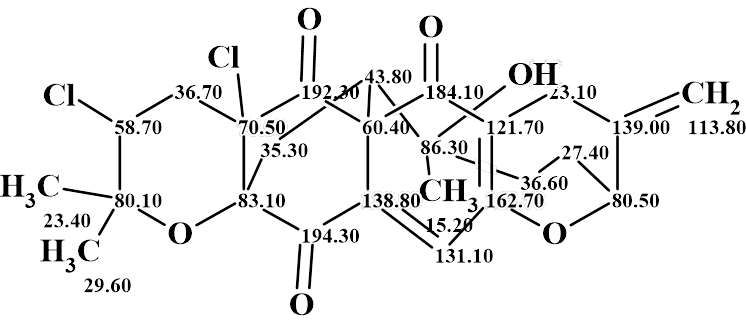
Thus, a complex and unusual structure of Canumycin C has been determined fully automatically in 12 minutes. The structure with automatically assigned chemical shifts is shown below:
References
- Hao, C. Qu, Y. Deng, L. Guo, T. Jin, M. Xu, P. Wang, W. Guo, L. Kou, S. Zhang, G. Hou, Z. Xie. (2025). Canumycins A–E, Macrocyclic Napyradiomycins from a Marine-Derived Streptomyces canus. Org. Lett., 27(22), 5555-5559. DOI: 10.1021/acs.orglett.5c00472
