February 1, 2017
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Sophaline C
The seeds and roots of the Sophora alopecuroides L plant, found in western and central Asia, have been used in traditional Chinese medicine for the treatment of various ailments. Previous investigations have found that matrine-type alkaloids and flavonoids are the main constituents of this plant. These have unique 6/6/6/6 tetracyclic skeletons. Their complex 3D structure and their biological activity have classed them as targets for both pharmacologists and synthetic chemists.
Just recently (2017) Y.-B. Zhang and co-workers [1] reported four novel matrine-based alkaloids, sophalines A-D. Their structures were elucidated using spectroscopic methods and single-crystal X-ray diffraction. Two of them (C and D) are a pair of stereoisomeric matrine-acetophenone alkaloids with an unprecedented skeleton.
We used the Spectroscopic data from [1] for Sophaline C (1) to challenge ACD/Structure Elucidator.
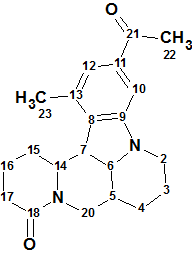
1
Sophaline C (1) was isolated as colorless crystals from CH3OH. The molecular formula of 1 was determined to be C21H26N2O2 on the basis of its high-resolution MS ion peak at m/z 339.2065 [M + H]+ (calcd for C21H27N2O2 339.2067). Its UV spectrum exhibited absorption maxima at 206 and 250 nm and the IR spectrum displayed characteristic absorptions for carbonyl bonds (1735 and 1628 cm-1) and an aromatic ring (1567 and 1444 cm-1). The HSQC, 1H –1H COSY, and HMBC data reported in [1] are summarized in Table 1.
Table 1. Sophaline C. NMR spectroscopic data.
| Label | δC | δC calc* | XHn | δH | M,J | COSY | C HMBC |
| C 2 | 45.200 | 43.570 | CH2 | 3.810 | u | ||
| C 2 | 45.200 | 43.570 | CH2 | 2.990 | u | 1.62 | C 9 |
| C 3 | 23.800 | 26.410 | CH2 | 1.620 | u | 1.32, 2.99 | |
| C 3 | 23.800 | 26.410 | CH2 | 1.620 | u | ||
| C 4 | 30.000 | 28.100 | CH2 | 1.880 | u | ||
| C 4 | 30.000 | 28.100 | CH2 | 1.320 | u | 1.62, 1.93 | C 20 |
| C 5 | 31.600 | 36.000 | CH | 1.930 | u | 1.32, 3.24, 3.38 | |
| C 6 | 68.100 | 63.470 | CH | 3.240 | u | 1.93, 3.52 | C 8, C 9 |
| C 7 | 49.500 | 44.580 | CH | 3.520 | u | 3.24, 4.04 | C 5, C 8, C 15 |
| C 8 | 135.300 | 134.070 | C | ||||
| C 9 | 154.100 | 151.280 | C | ||||
| C 10 | 104.400 | 106.390 | CH | 6.880 | S | C 8, C 12, C 21 | |
| C 11 | 138.800 | 135.760 | C | ||||
| C 12 | 123.400 | 123.200 | CH | 7.130 | S | ||
| C 13 | 137.000 | 134.510 | C | ||||
| C 14 | 55.100 | 54.830 | CH | 4.040 | u | 2.25, 3.52 | |
| C 15 | 31.100 | 28.890 | CH2 | 2.250 | u | 1.77, 4.04 | |
| C 15 | 31.100 | 28.890 | CH2 | 2.120 | u | ||
| C 16 | 19.200 | 19.710 | CH2 | 1.770 | u | 2.25, 2.41 | |
| C 16 | 19.200 | 19.710 | CH2 | 1.930 | u | ||
| C 17 | 33.300 | 30.890 | CH2 | 2.410 | u | 1.77 | C 18 |
| C 17 | 33.300 | 30.890 | CH2 | 2.410 | u | ||
| C 18 | 173.400 | 171.310 | C | ||||
| C 20 | 47.200 | 42.080 | CH2 | 3.380 | u | 1.93 | C 18 |
| C 21 | 200.600 | 197.590 | C | ||||
| C 22 | 26.800 | 26.570 | CH3 | 2.540 | S | C 21 | |
| C 23 | 21.800 | 19.660 | CH3 | 2.390 | S | C 12, C 13 |
13C chemical shift prediction was performed using the HOSE code based approach [2].
Figure 1 shows the slightly edited Molecular Connectivity Diagram (MCD) produced by Structure Elucidator.
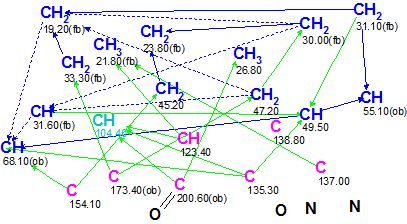
Figure 1. Sophaline C. Molecular Connectivity Diagram.
MCD overview. The molecular formula of Sophaline (C21H26N2O2) shows that the compound is hydrogen-rich and this is the reason why the COSY spectrum (blue lines) contains many correlations. Some COSY correlations are ambiguous (drawn by blue dotted lines) because carbon atoms C6 and C16 have hydrogens attached whose signals at 1.93 ppm are overlapping. Presence of ambiguous correlations usually leads to an increase in generation time, therefore some MCD edits are desirable in such situations. The methyne atomat 104.40 ppm is marked by light blue color. This means that its hybridization is automatically set as “not sp (sp3 or sp2)”: the atom can either belong to a C=C double bond or be connected to two heteroatoms being in sp3 state of hybridization. An evident ketone carbonyl bond was drawn manually for the carbon C 200.6, and label “ob” was assigned to C 55.10, 68.10, and C 173.40 (obligatory connection to any heteroatom).
Strict structure generation together with 13C chemical shift prediction and structural filtering was run, producing 59 structures. The following results were obtained: k = 2905 → 71 → 59, tg = 59 sec. 13C chemical shift prediction was applied to all items of the output file using additive rules, neural networks and fragmental (HOSE code-based) approaches [2], and average chemical shift deviations (dI, dN and dA) were calculated. All 59 structures were then ranked in increasing order of dA values. The four top-ranked structures are presented in Figure 2.
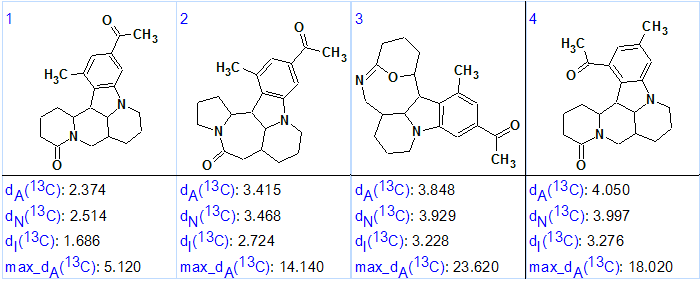
Figure 2. Sophaline C. Four top ranked structures of the output file.
Figure 2 shows that the first ranked structure (#1) is identical to structure of Sophaline C 1 that was confirmed by the single crystal X-ray analysis. Both the average deviations and the maximum deviation max_dA(13C) calculated for structure #1 indicate its validity. Note that structure #4, which differs from 1 by exchanged substituents on the aromatic ring, was eliminated by the ranking procedure with mean deviation of 4.05 ppm and maximum deviation of 18.02 ppm versus 2.374 and 5.120 ppm for structure #1. The structure of Sophaline C, along with automatically assigned 13C chemical shifts by Structure Elucidator, is shown below:
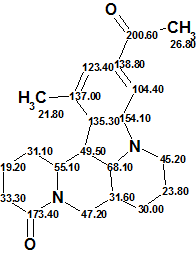
In conclusion a structure characterized by an unprecedented skeleton was determined with the help of ACD/Structure Elucidator automatically in one minute.
References
- Y.-B. Zhang, X.-L. Zhang, N.-H. Chen, Z.-N. Wu, W.-C. Ye,Y.-L. Li, G.-C. Wang. (2017). Four Matrine-Based Alkaloids with Antiviral Activities against HBV from the Seeds of Sophora alopecuroides. Org. Lett., 19, 424-427.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.


