March 1, 2022
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Lobocrassin A
Cembrane-type metabolites are among the largest group of diterpenoids isolated from octocorals. A rich source of such compounds has been found to be Lobophytum crassum, a soft coral from the family of Alcyoniidae. This coral has been studied by Kao et al [1], and five new cembrane derivatives have been isolated. Amongst them is lobocrassin A (1) which was identified based on MS and 1D and 2D NMR spectra.
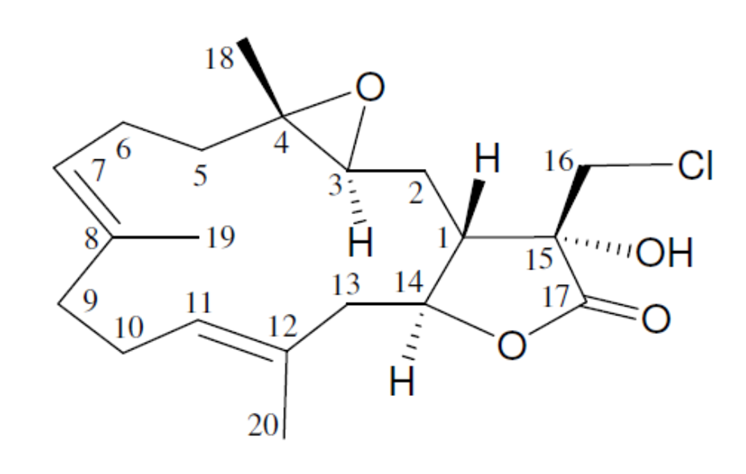
1
Lobocrassin A (1) was isolated as a colorless oil, and the molecular formula for this compound was determined to be C20H29ClO4 (six units of unsaturation) using HR-ESI-MS (C20H2935ClO4 + H, m/z 369.1830, calculated 369.1833). Comparison of the 13C NMR and DEPT data with the molecular formula indicated that there must be an exchangeable proton, which required the presence of a hydroxy group. This assumption was supported by a broad absorption in the IR spectrum at 3385 cm–1. The IR spectrum also showed a strong band at 1778 cm–1, consistent with the presence of a g-lactone moiety.
The spectroscopic data presented in [1] were used by us to challenge ACD/Structure Elucidator (ACD/SE). In so doing, we only used 1D 13C and 1H and 2D HSQC and HMBC data (Table 1).
Table 1. NMR spectroscopic data.
| C/X Label | δC | δC calc (HOSE) | XHn | δH | M | H to C HMBC |
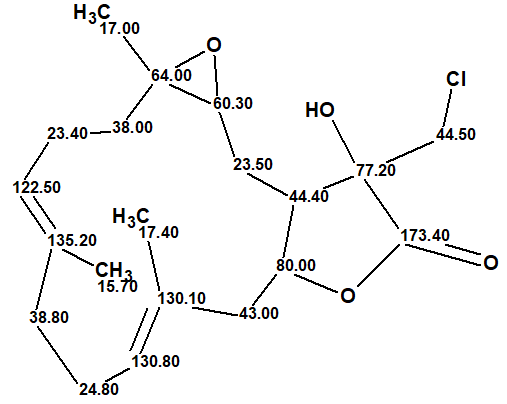
| C 1 | 44.4 | 44.52 | CH | 2.76 | u | C 2, C 13, C 16, C 14 |
| C 2 | 23.5 | 27.4 | CH2 | 1.68 | u | |
| C 2 | 23.5 | 27.4 | CH2 | 2.14 | u | C 1, C 3, C 4, C 15, C 14 |
| C 3 | 60.3 | 60.69 | CH | 2.86 | u | C 2 |
| C 4 | 64 | 61.11 | C | |||
| C 5 | 38 | 36.42 | CH2 | 2.07 | u | C 3, C 4, C 7 |
| C 5 | 38 | 36.42 | CH2 | 1.29 | u | |
| C 6 | 23.4 | 23.74 | CH2 | 2.24 | u | C 4, C 7, C 8 |
| C 6 | 23.4 | 23.74 | CH2 | 2.09 | u | |
| C 7 | 122.5 | 125.9 | CH | 5.07 | u | C 19, C 6, C 9 |
| C 8 | 135.2 | 134.7 | C | |||
| C 9 | 38.8 | 38.75 | CH2 | 2.26 | u | C 19, C 10, C 7, C 11, C 8 |
| C 10 | 24.8 | 24.85 | CH2 | 2.21 | u | |
| C 10 | 24.8 | 24.85 | CH2 | 2.32 | u | C 9, C 12, C 11, C 8 |
| C 11 | 130.8 | 128.62 | CH | 5.23 | u | C 20, C 10, C 9 |
| C 12 | 130.1 | 130.68 | C | |||
| C 13 | 43 | 45.7 | CH2 | 2.52 | u | |
| C 13 | 43 | 45.7 | CH2 | 2.67 | u | C 1, C 14, C 12, C 11 |
| C 14 | 80 | 76.13 | CH | 4.66 | u | C 12 |
| C 15 | 77.2 | 76.02 | C | |||
| C 16 | 44.5 | 46.08 | CH2 | 3.53 | u | |
| C 16 | 44.5 | 46.08 | CH2 | 3.79 | u | C 1, C 15, C 17 |
| C 17 | 173.4 | 176.24 | C | |||
| C 18 | 17 | 18.7 | CH3 | 1.34 | s | C 5, C 3, C 4 |
| C 19 | 15.7 | 15.9 | CH3 | 1.6 | s | C 9, C 7, C 8 |
| C 20 | 17.4 | 17.55 | CH3 | 1.74 | s | C 13, C 12, C 11 |
| O 1 | OH | 4.03 | u | C 1, C 16, C 15, C 17 |
The automatically generated Molecular Connectivity Diagram (MCD) is shown in Figure 1.
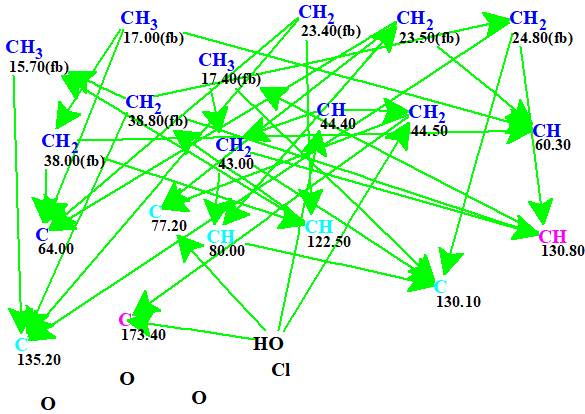
Figure 1. Automatically generated Molecular Connectivity Diagram. The hybridizations of carbon atoms are marked by the corresponding colors: sp2 – magenta, sp3 – blue, not sp – light blue. Labels “ob” and “fb” are set by the program to carbon atoms for which neighboring with heteroatom is either obligatory (ob) or forbidden (fb).
No manual edits were made in the MCD, and strict structure generation accompanied with 13C chemical shift prediction using incremental approach was initiated. Result: k=12 → (spectral filtering) → 3 → (duplicate removal) → 3, tg = 2 s. Afterwards 13C chemical shift prediction was carried out using the HOSE code-based method and neural networks. The average deviations of 13C chemical shifts determined by these methods are denoted as dA and dN correspondingly. Finally, the output file was ranked in increasing order of dA values. The ranked structures are shown in Figure 2.
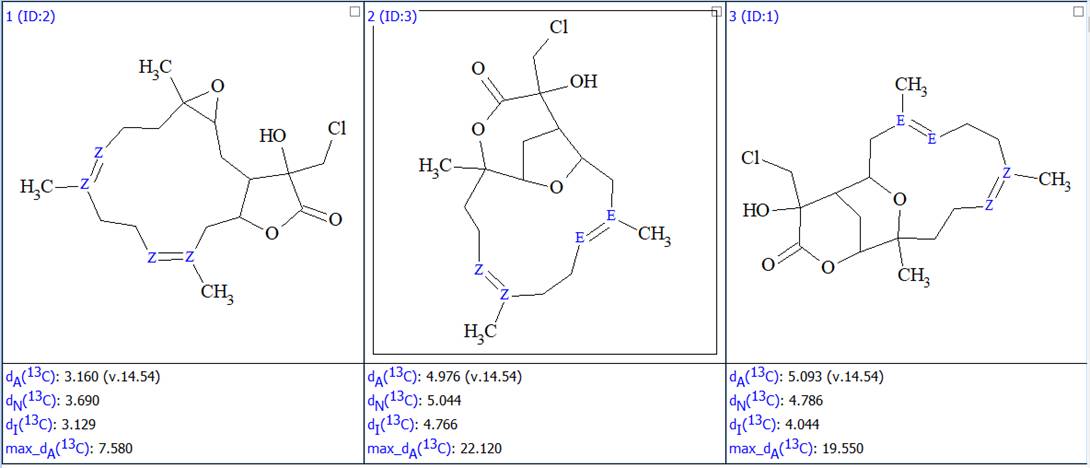
Figure 2. The ranked output file showing the 3 generated structures.
We see that the best structure coincides with the structure of lobocrassin A determined by the authors of the article [1]. However, structure #1 was drawn by the program without considering the possible configurations of the double bonds C=C. Since HOSE-codes based 13C chemical shift prediction is sensitive to the structure configuration we used the following option in the Structure menu (Figure 3):
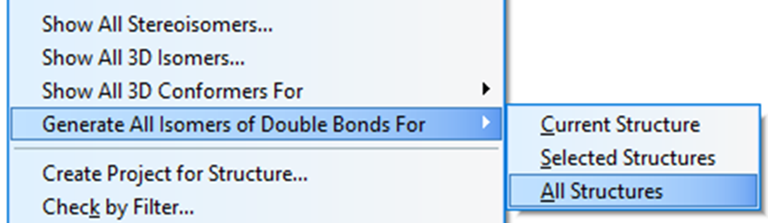
Figure 3. Menu option Structure->Generate All Isomers of Double Bonds->All Structures
As a result, 8 structures were generated. After recalculating the 13C chemical shifts and ranking the structures the resulting 8 structures together with their scores are shown in Figure 4:
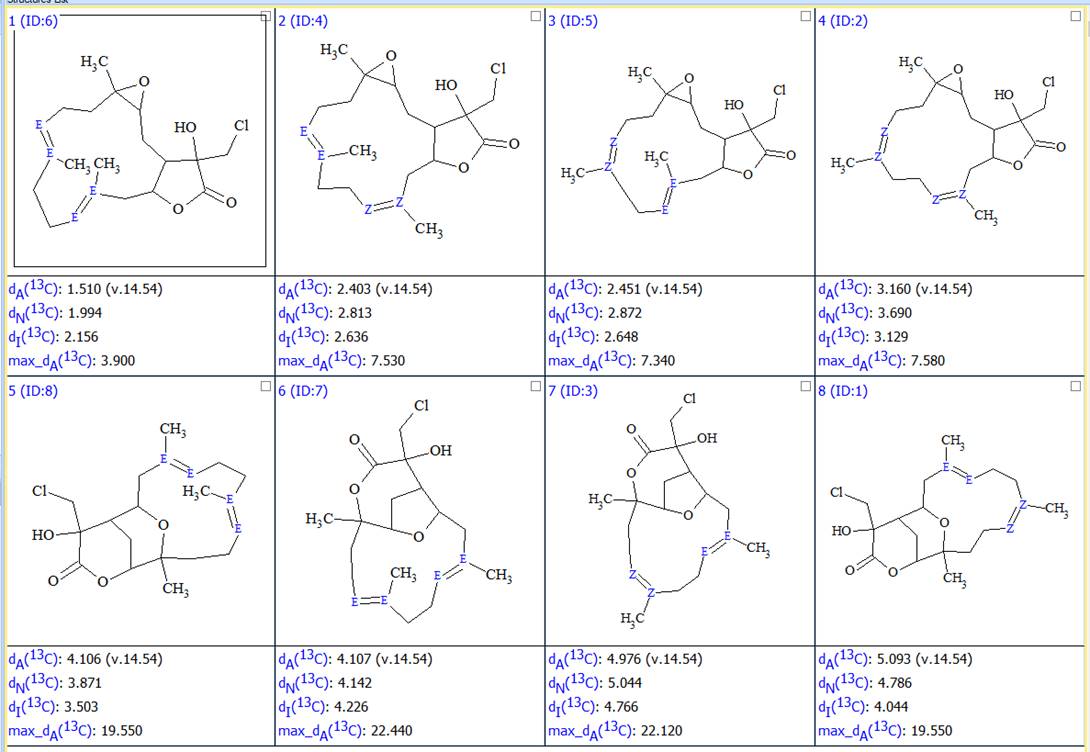
Figure 4. All possible configurations of structures presented in Figure 2.
Furthermore the duplicate structures were removed and the final output file optimized, bythe double bond configurations in cyclic structures, was obtained (Figure 5).
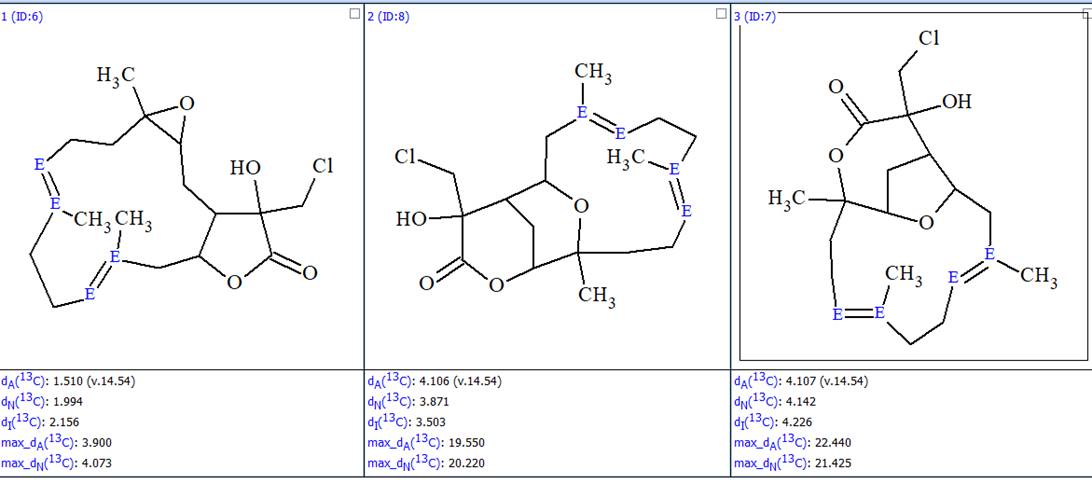
Figure 5. The ranked output file whose structures are optimized to the most probable configurations.
In this way, the correct structure and its most probable E/Z configuration of lobocrassin A was determined by ACD/SE in fully automatic mode. The structure together with the 13C chemical shift assignment is shown below.