December 13, 2023
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Pseudochelin A
Sonnenschein and collaborators [1] investigated the fungus Pseudoalteromonas piscicida S2040. The crude extract of the culture of Pseudoalteromonas piscicida S2040 was fractionated by reversed phase solid phase extraction (SPE) before further purification by reversed phase HPLC to yield pseudochelin A (1) – a new siderophore containing a 4,5-dihydroimidazole moiety.
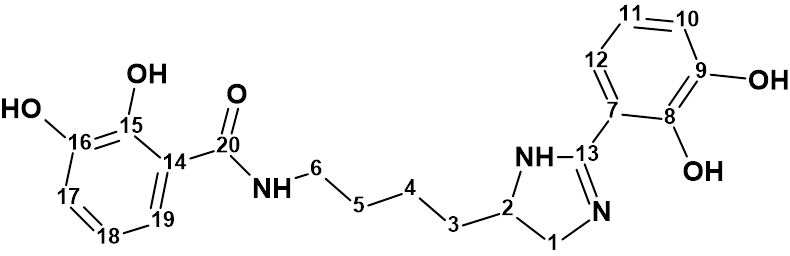
1
The structure of pseudochelin A was determined by a combined employment of HR-ESI-MS and 1D and 2D NMR data. Pseudochelin A showed a high resolution ESI-MS peak of m/z 386.172 [M+H]+ (Δ 0.5 ppm) from calculated for C20H24N3O5 , and its molecular formula was determined as C20H23N3O5. To challenge Structure Elucidator suite, the 1H, 13C , HSQS, HMBC and COSY data (Table 1) published in article [1] were entered into the program.
Table 1. NMR spectroscopic data for pseudochelin A[1]
| C Label | δC | δC calc (HOSE) | XHn | δH | COSY | H to C HMBC |
| C 1 | 49.1 | 50.6 | CH2 | 4.12 | C 3, C 2, C 13 | |
| C 1 | 49.1 | 50.6 | CH2 | 3.71 | 4.36 | |
| C 2 | 56.7 | 58.1 | CH | 4.36 | 1.86, 3.71 | C 1, C 13 |
| C 3 | 34.2 | 35.6 | CH2 | 1.86 | 1.52, 4.36 | C 2 |
| C 3 | 34.2 | 35.6 | CH2 | 1.77 | ||
| C 4 | 21.5 | 22.96 | CH2 | 1.52 | 1.86 | C 5, C 6 |
| C 4 | 21.5 | 22.96 | CH2 | 1.48 | 1.7 | |
| C 5 | 28.8 | 30.2 | CH2 | 1.7 | 1.48, 3.45 | C 4, C 3, C 6 |
| C 6 | 38.5 | 40 | CH2 | 3.45 | 1.7 | C 4, C 5, C 20 |
| C 7 | 107.8 | 109.3 | C | |||
| C 8 | 147.5 | 149 | C | |||
| C 9 | 146.1 | 147.6 | C | |||
| C 10 | 119.9 | 121.3 | CH | 7.08 | 6.84 | C 12, C 9, C 8 |
| C 11 | 119.8 | 121.3 | CH | 6.84 | 7.08, 7.14 | C 7, C 9 |
| C 12 | 118.7 | 120.1 | CH | 7.14 | 6.84 | C 11, C 10, C 8, C 13 |
| C 13 | 163 | 164.5 | C | |||
| C 14 | 115.4 | 116.8 | C | |||
| C 15 | 148.7 | 150.2 | C | |||
| C 16 | 145.9 | 147.4 | C | |||
| C 17 | 118.1 | 119.6 | CH | 6.91 | 6.7 | C 19, C 16, C 15 |
| C 18 | 118.2 | 119.6 | CH | 6.7 | 6.91, 7.18 | C 14, C 16 |
| C 19 | 117.2 | 118.6 | CH | 7.18 | 6.7 | C 17, C 18, C 15, C 20 |
| C 20 | 170.1 | 171.5 | C |
A Molecular Connectivity Diagram (MCD) automatically created by the software is presented in Figure 1.
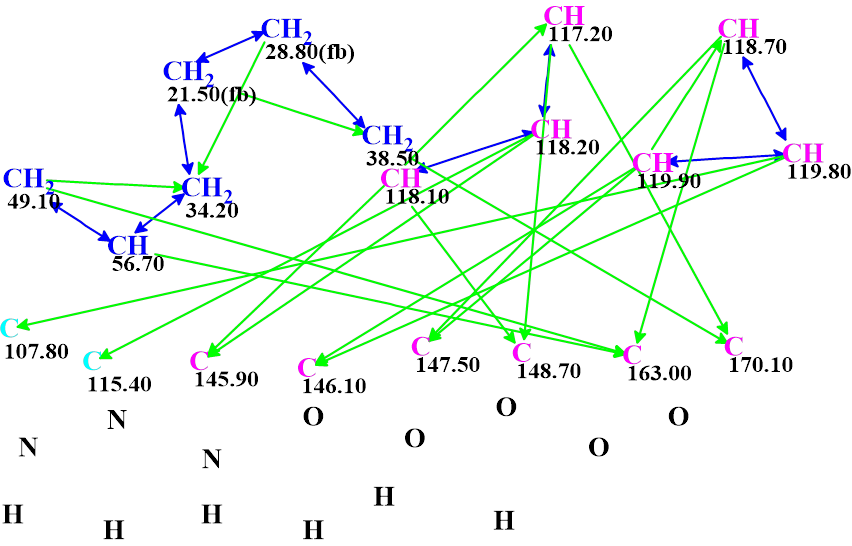
Figure 1. The molecular connectivity diagram for pseudochelin A. Hybridizations of carbon atoms are marked by corresponding colors: sp2 – violet, sp3 – blue, not sp (sp3 or sp2) – light blue. The “fb” labels are set by the program to carbon atoms for which neighboring with heteroatom is forbidden (fb). The HMBC connectivities are marked by green arrows, while COSY connectivities – by blue arrows.
No MCD edits were made. Checking the MCD for presence of contradictions in the combined HMBC and COSY data showed that the data were consistent, therefore strict structure generation followed by 13C chemical shift prediction and structure filtering was initiated. Results: k = 212 → (structural filtering) → 36 → (duplicate removal) → 36, tg = 1.5 s.
Then 13C chemical shift prediction was carried out for all 36 structures using the three empirical methods (HOSE code based, neural networks, incremental approach) and the output file was ranked in increasing order of average chemical shift deviations.
The nine top ranked structures are shown in Figure 2.
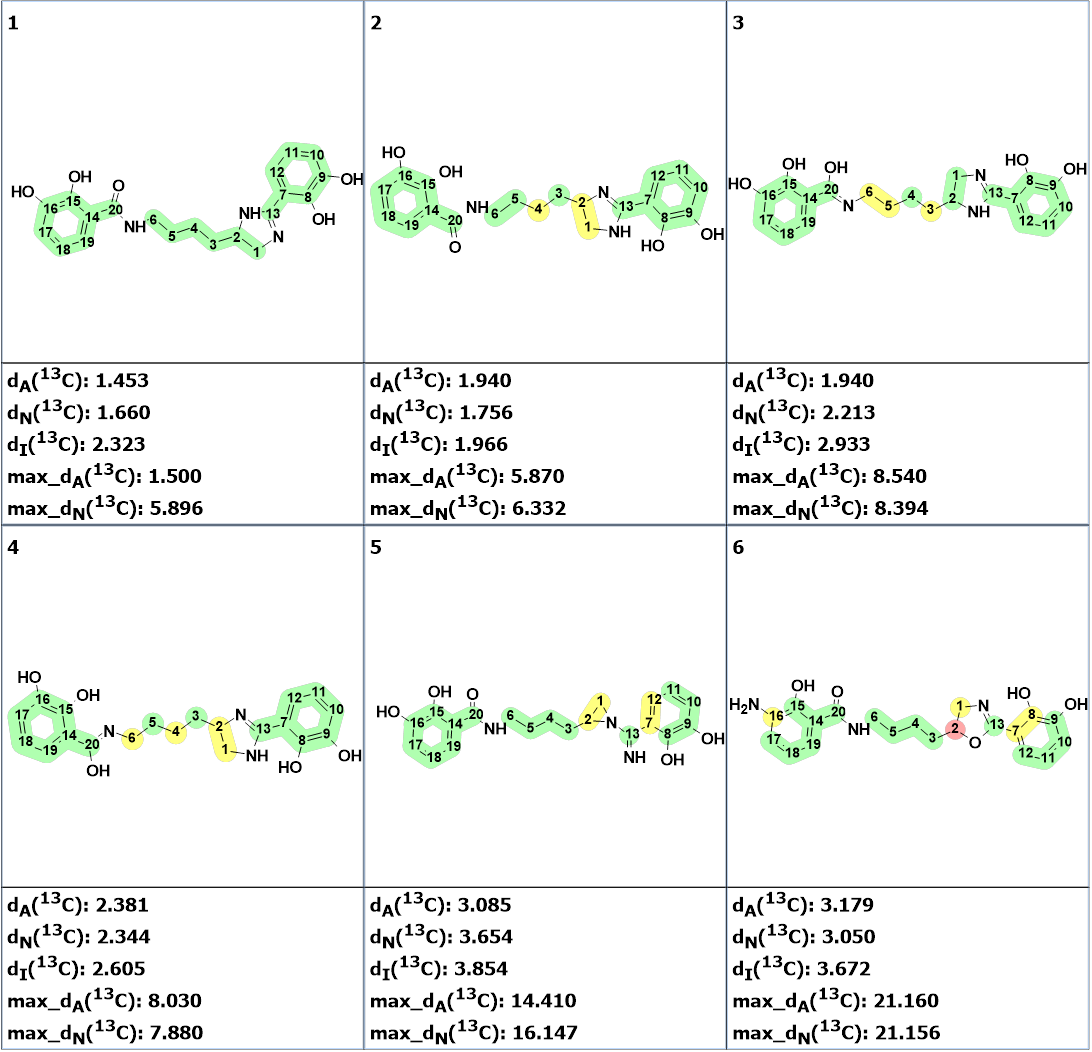
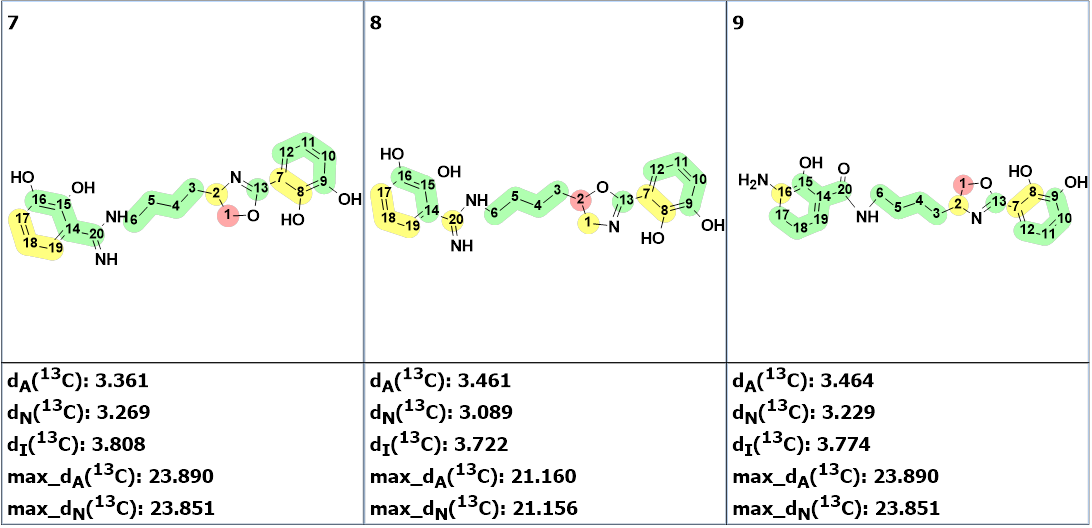
Figure 2. The nine top ranked generated structures. The 13C chemical shift prediction was carried out using the HOSE code-based method, the neural networks, and the incremental approach. Average deviations of 13C chemical shifts determined by these methods are denoted as dA, dN and dI correspondingly. Each atom is colored to mark a difference between its experimental and calculated 13C chemical shifts. The green color represents a difference between 0 to 3 ppm, yellow was >3 to 15 ppm, red > 15 ppm.
Figure 2 shows that structure #1 is identical to the structure of pseudochelin A determined by the authors [1]. All atoms of this structure are green what means that deviations calculated for each carbon atom, using HOSE code approach, are not exceeding 3 ppm (see Table 1). Our experience shows that a fully “green structure” is usually a sign of its validity. At the same time, values of average and maximum deviations obtained for structures #2-#4 can be considered acceptable. Therefore, empirical DP4 probabilities were calculated for structures #1- #4, assuming that all these structures are equiprobable. The results are shown in Figure 3.
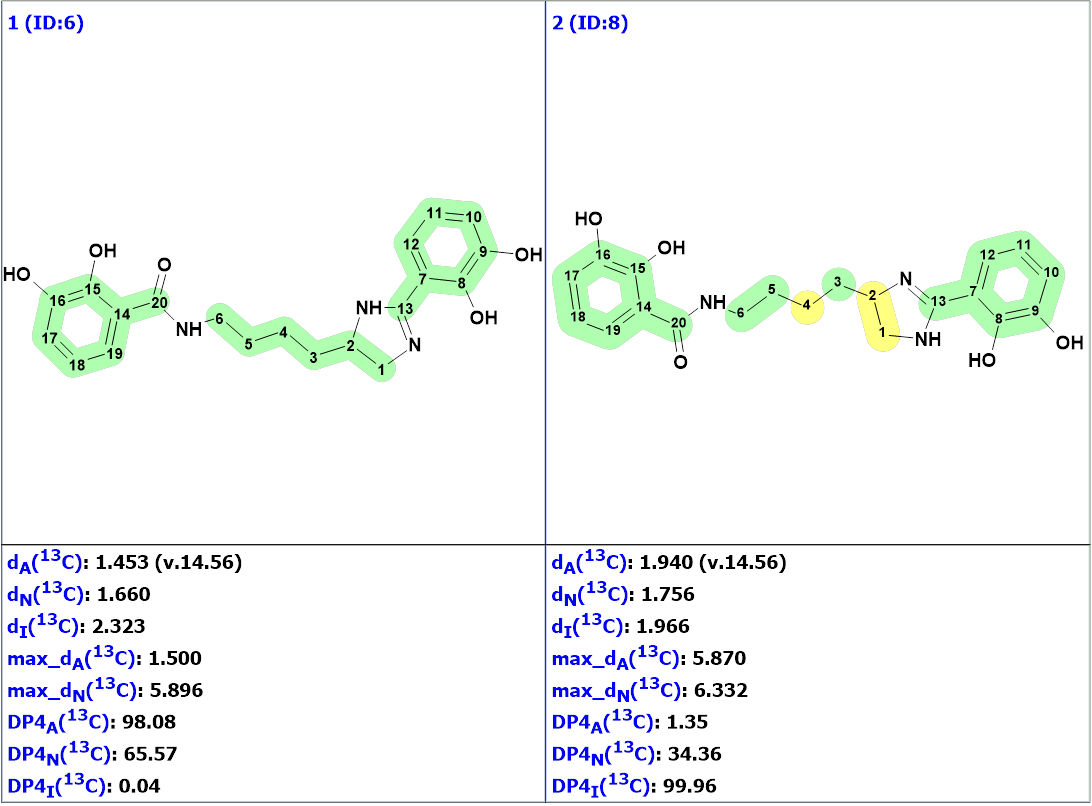
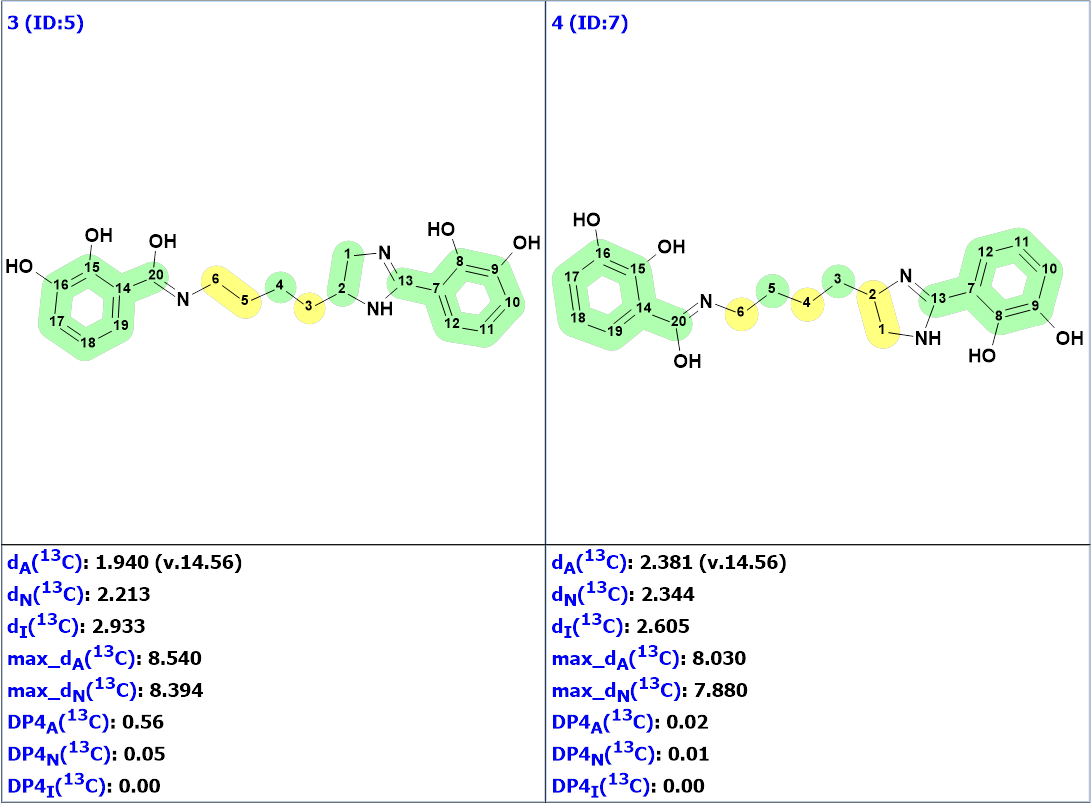
Figure 3. DP4 probabilities calculated for structures #1 – #4.
We see that structures #3 and #4 can be confidently rejected, while structure #1 is the most probable one. The validity of this structure is supported by the fact that the incremental approach which produced DP4I(13C)=99.96 for structure #2 is characterized by the lowest accuracy in comparison to the other two methods of chemical shift prediction implemented in Structure Elucidator. Meanwhile structure #2 is a tautomer of structure #1. According to the methodology of evaluating structures generated by CASE [2-3], it would be interesting to perform DFT based chemical shift prediction for both tautomers. This was done by authors [1] who found that the correlation of 13C experimental and calculated chemical shift values did not show any significant difference between the two structures based on the regression coefficient values.
Structure #1 with the assigned 13C chemical shifts is shown below:
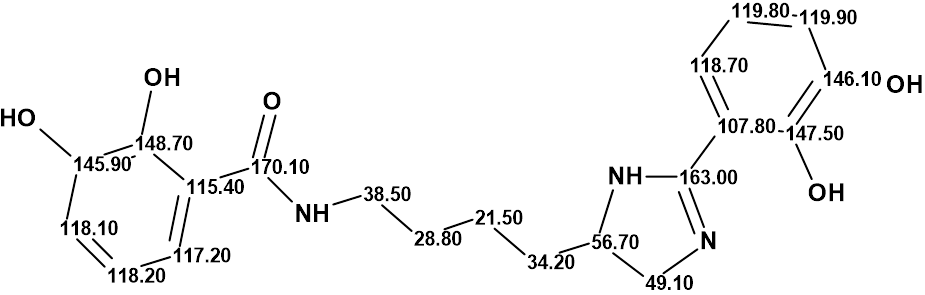
Thus, a challenging pseudochelin A structure containing eight heteroatoms (three atoms of nitrogen and fife atoms of oxygen) was elucidated by Structure Elucidator fully automatically in 1.5 seconds.
References
- Sonnenschein, E. C.; Stierhof, M.; Goralczyk, S.; Vabre, F. M.; Pellissier, L.; Hanssen, K. Ø.; de la Cruz, M.; Díaz, C.; de Witte, P.; Copmans, D.; Andersen, J. H.; Hansen, E.; Kristoffersen, V.; Tormo, J. R.; Ebel, R.; Milne, B. F.; Deng, H.; Gram, L.; Jaspars, M.; Tabudravu, J. N. (2017). Pseudochelin A, a siderophore of Pseudoalteromonas piscicida S2040. Tetrahedron, 73, (18), 2633-2637.
- Buevich, A. V.; Elyashberg M. E. (2016). Synergistic combination of CASE algorithms and DFT chemical shift predictions: a powerful approach for structure elucidation, verification and revision. J. Nat. Prod., 79(12), 3105–3116.
- Buevich, A.V.; Elyashberg M. E. (2018). Towards unbiased and more versatile NMR-based structure elucidation: A powerful combination of CASE algorithms and DFT calculations. Magn. Reson. Chem., 56, 493–504.


