October 17, 2023
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Nordine
Sesquiterpenoids are a well-known class of compounds with an important role in biological systems. They are found in many plants and microorganisms. They are known to have complex structures and exhibit various biological activities. The macrocyclic humulene-type compounds are a type of sesquiterpenoids with known structural diversity which includes bicyclic and tricyclic moieties and different functional groups.
Due to the wide variability of these compounds, it is often challenging to determine the exact structure and avoid mistakes. However, this is important in order to properly understand the biological effects these compounds have. Different approaches have been used for this, including computational chemistry, structural derivatizations, NMR spectroscopic methods, X-ray crystallography techniques, and biosynthetic studies.
In 2012 Husain and co-workers [1] reported a new macrocyclic humulene-type sesquiterpenoid called nordine, which was obtained from the roots of Anaxagorea javanica Blume.
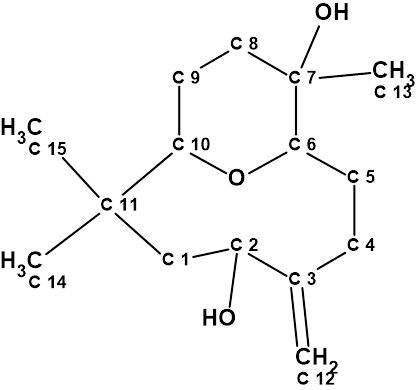
1
Nordine was proposed to contain an ether-bridged bicyclic ring between C-10 and C-6 and bears a hydroxy group at C-2. Its structure determination was based on the analysis of 1D and 2D NMR, IR, and HR-EI-MS data.
In 2022, Tavares and co-workers [2] also isolated Nordine. The molecular formula was determined as C15H26O3 by HR-ESI-MS. The 1D and 2D NMR data for 1 were identical to those described for Nordine in [1]. However, the authors [2] realized that the values of chemical shifts for the carbons C-6 and C-7 may indicate another ether fusion, and they suggested that the real structure of Nordine was 2:
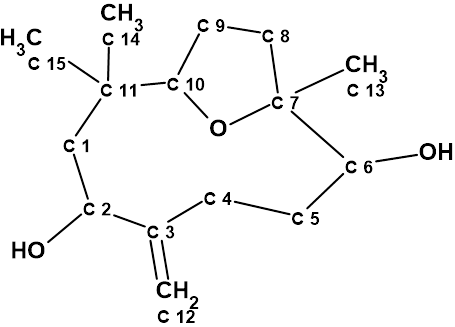
2
Therefore structure 1 was revised to 2, and the revision was confirmed by literature data, obtaining the diacetylated derivative by X-ray crystallography.
It is known that most frequently erroneous structures are deduced when the unknown compounds are complex and hydrogen deficient. The structure of Nordine is relatively small with a good amount of hydrogen atoms. So, it was interesting to learn what led the authors of [1] to the incorrect structure.
With this in mind, we entered the molecular formula and NMR spectroscopic data of Nordine (Table 1) in ACD/Structure Elucidator.
Table 1. NMR spectroscopic data of Nordine.
| Label | δC | XHn | δH | M(H) | H to C HMBC |
| C 1 | 46.6 | CH2 | 2.13 | u | C 15, C 14, C 11, C 2, C 10, C 3 |
| C 1 | 46.6 | CH2 | 1.51 | u | |
| C 2 | 69.5 | CH | 4.42 | u | C 4, C 11, C 1, C 12, C 3 |
| C 3 | 157.1 | C | |||
| C 4 | 32.4 | CH2 | 2.4 | u | |
| C 4 | 32.4 | CH2 | 2.52 | u | C 5, C 2, C 6, C 12, C 3 |
| C 5 | 30.2 | CH2 | 1.55 | u | |
| C 5 | 30.2 | CH2 | 2.22 | u | C 4, C 6, C 7, C 3 |
| C 6 | 73.7 | CH | 3.34 | u | C 13, C 5, C 4, C 8, C 7 |
| C 7 | 84.4 | C | |||
| C 8 | 35.5 | CH2 | 1.55 | u | |
| C 8 | 35.5 | CH2 | 2.08 | u | C 9, C 6, C 7, C 10 |
| C 9 | 26.6 | CH2 | 1.6 | u | |
| C 9 | 26.6 | CH2 | 1.68 | u | C 8, C 11, C 7, C 10 |
| C 10 | 87.8 | CH | 3.64 | u | C 15, C 9, C 1 |
| C 11 | 35.8 | C | |||
| C 12 | 112.6 | CH2 | 5.3 | u | C 4, C 2, C 3 |
| C 12 | 112.6 | CH2 | 5.01 | u | |
| C 13 | 20.5 | CH3 | 1.18 | s | C 8, C 6, C 7 |
| C 14 | 28.5 | CH3 | 0.78 | s | C 15, C 11, C 1, C 10 |
| C 15 | 25.8 | CH3 | 1.2 | s | C 14, C 11, C 1, C 10 |
The Molecular Connectivity Diagram (MCD) was created automatically by the program (Figure 1).
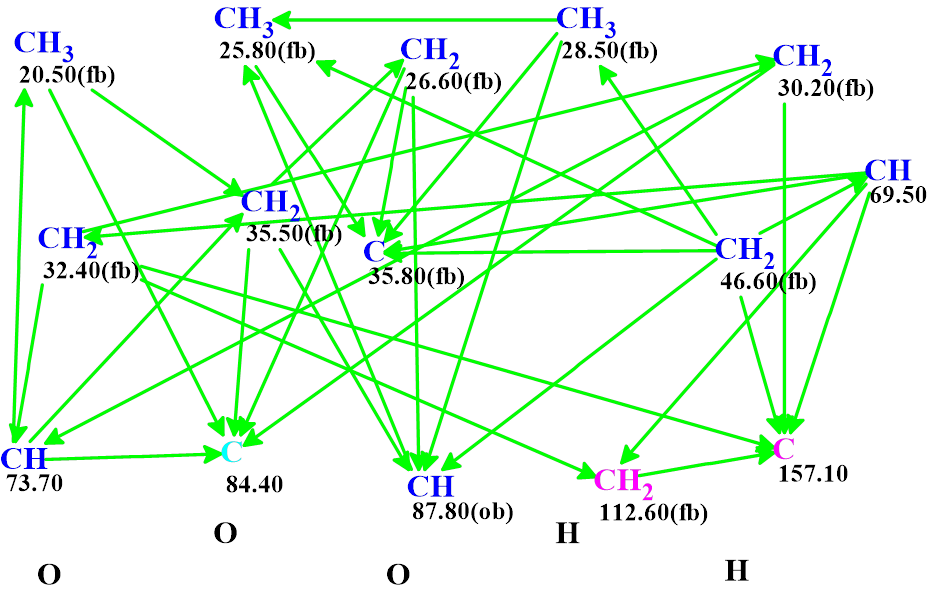
Figure 1. The Molecular Connectivity Diagram (MCD) of Nordine. The hybridizations of carbon atoms are marked by corresponding colors: sp2 – violet, sp3 – blue, not sp (sp3 or sp2) – light blue. The labels “ob” and “fb” are set by the program to carbon atoms for which neighboring with heteroatom is either obligatory (ob) or forbidden (fb). The HMBC connectivities are marked by green arrows.
Figure 1 shows that the network of many HMBC correlations (50 arrows) connects all carbon atoms. Checking the MCD for the presence of contradictions (nonstandard correlations with a length > 3 chemical bonds) was completed with the program message informing that the HMBC data are consistent, and strict structure generation accompanied with 13C chemical shift prediction and structural filtering was initiated. As a result, 6 structures were generated in 0.5 s. 13C chemical shift prediction was carried out for all structures using the three methods implemented into ACD/Structure Elucidator, and the structures were ranked in increasing order of average deviations of calculated chemical shifts from experimental ones. The ranked output file is presented in Figure 2.
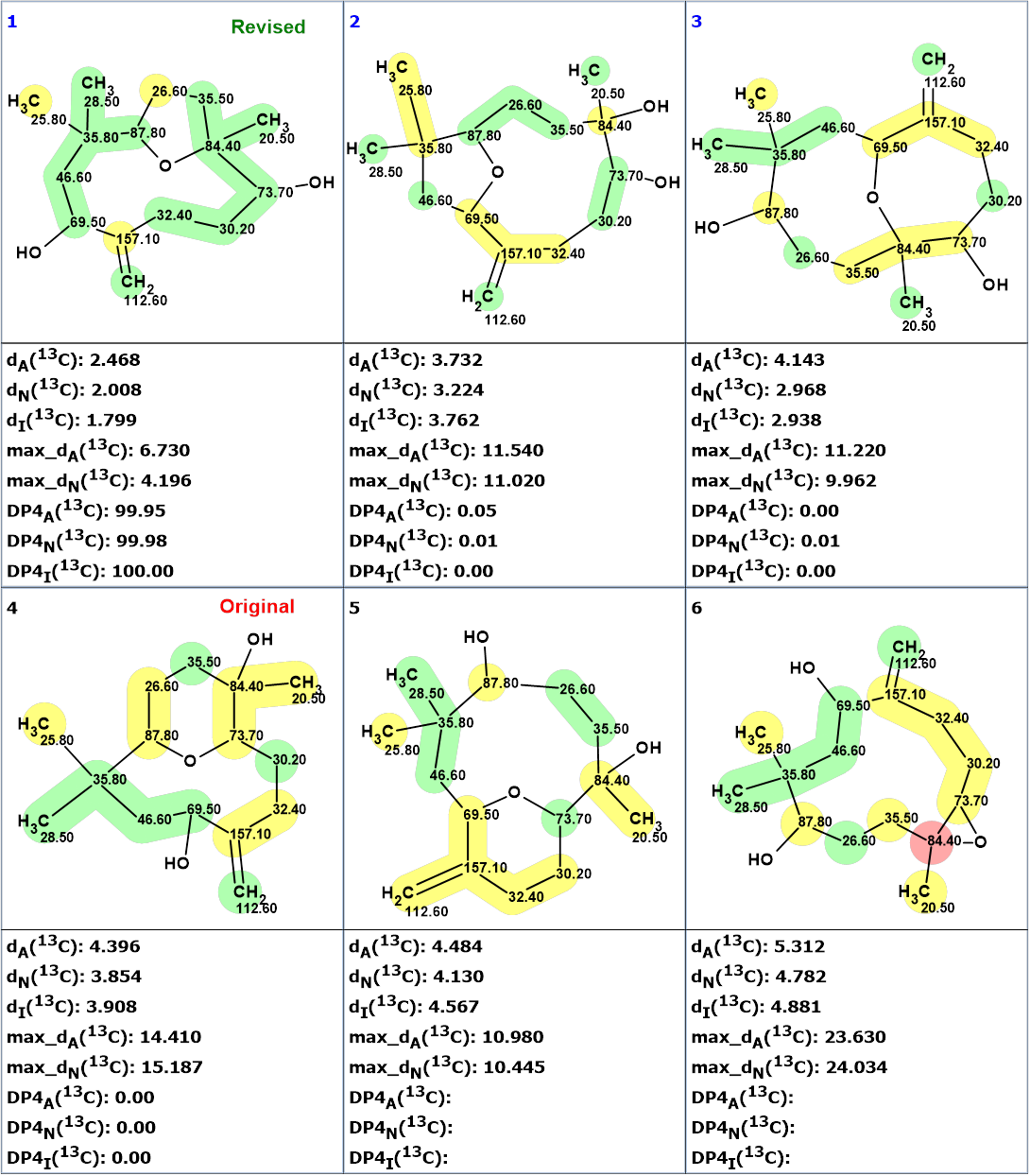
Figure 2. The ranked output file. 13C chemical shift prediction was carried out using the HOSE code-based method, the neural networks, and the incremental approach. Average deviations of 13C chemical shifts determined by these methods are denoted as dA, dN and dI correspondingly. Each atom is colored to mark a difference between its experimental and calculated 13C chemical shifts. The green color represents a difference between 0 to 3 ppm, yellow was >3 to 15 ppm, red > 15 ppm.
Figure 2 shows that the revised structure 2 was ranked as first while the original structure 1 was placed in the 4th position. The calculation of empirical DP4 probabilities for structures #1–#4 confirmed the validity of the revised structure 2.
Note that all generated structures fully satisfy all HMBC correlations, and their probabilities can be evaluated only on the basis of spectrum prediction. We suggest that in this case, the reason for the wrong structure determination was the great number of correlations that the authors [1] failed to analyze simultaneously. The strength of the CASE approach is in the fact that the program keeps all correlations in memory and performs logical inference of all conceivable structures from the initial information. Obviously, if the authors [1] utilized ACD/SE or at least ACD/CH-Predictors during the structure elucidation, they would most probably have avoided the structure misassignment.
References
- Husain, K.; Zakaria, S. M.; Lajis, N. H.; Shaari, K.; Ismail, I. S.; Israf, D. A.; Paetz, C. (2012). Phytochem. Lett., 5, 788–792.
- R. S. de Andrade, K. A. Sales, L. S. Abreu, V. R. Campos, F. M. dos Santos Junior, R. Braz-Filho, M. T. Scotti, J. F. Tavares, M. S. da Silva. (2022). Structure Revision of the Sesquiterpene Nordine Based on NMR Spectroscopic Analysis and X‑ray Crystallography. J. Nat. Prod., 85, 2480−2483.


