April 9, 2024
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Janthinolide A
A new 2,5-piperazinedione derivative, janthinolide A (1), was obtained from the fermentation broths of the endophytic fungus Penicillium janthinellum [1]. The latter was isolated from a soft coral, Dendronephthya sp. The structure of janthinolide A was established using MS spectrometry in combination with 1D and 2D NMR data. The molecular formula of janthinolide A (1) was determined to be C23H32N2O7 on the basis of HR-FAB-MS (m/z 449.2281, calcd 449.2282). The structure of 1 together with 13C NMR chemical shift assignments as suggested by the authors [1] is presented below:

1
However, just recently the authors of [2] showed that the true structure of 1 is 2:
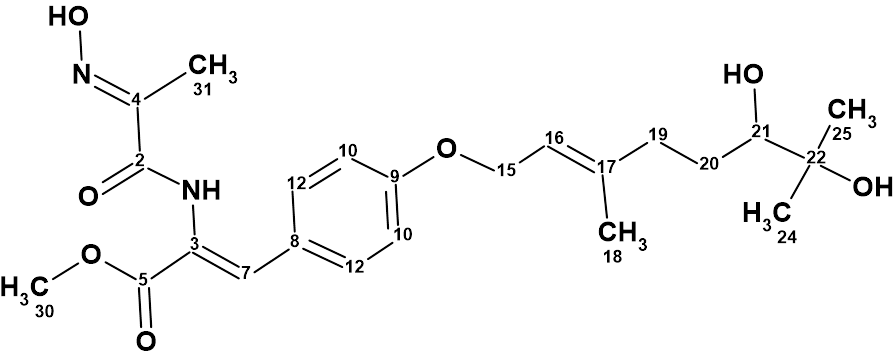
2
To revise structure 1, they utilized Structure Elucidator Suite combined with DFT based NMR chemical shift prediction. First, structure 1 was entered as a PS (Proposed Structure) for Structure Elucidator and 13C chemical shift prediction was carried out using neural networks and incremental approaches. Employment of the HOSE code based method of prediction was impossible for this unusual structure as it contains a radical. The result is shown in Figure 1.
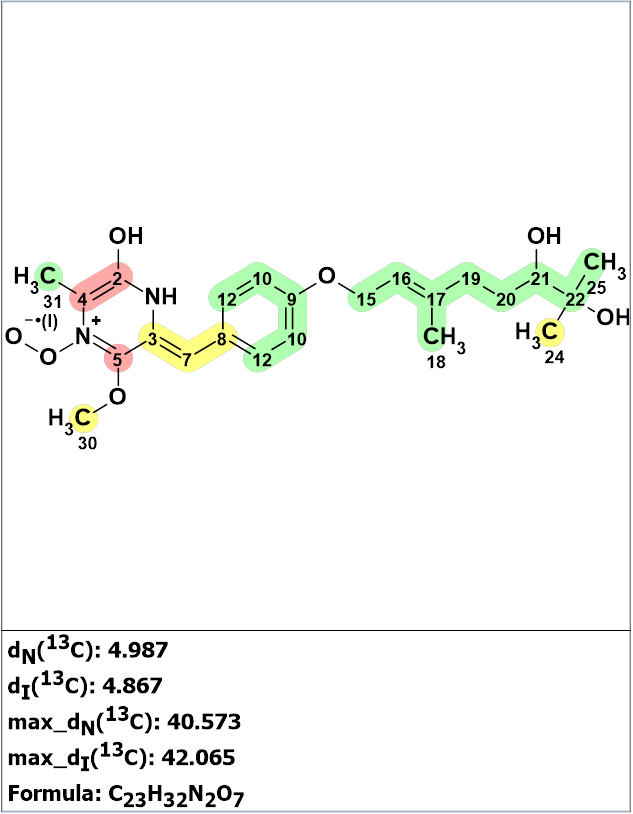
Figure 1. The structure of janthinolide A proposed by Xue et al [1] for which 13C chemical shift prediction was carried out using the neural networks and the incremental approach. Average deviations of 13C chemical shifts determined by these methods are denoted as dN and dI correspondingly. Each atom is colored to mark a difference between its experimental and calculated 13C chemical shifts. The green color represents a difference between 0 to 3 ppm, yellow was >3 to 15 ppm, red >15 ppm.
Figure 1 shows that the predicted chemical shifts were consistent with the dihydroxylated geranyl ether moiety (a “green fragment”), but major inconsistencies were revealed between the calculated and observed 13C NMR chemical shifts for the diketopiperazine-derived N-heterocycle. The average deviations of predicted 13C NMR chemical shifts from the experimental values are approximately 5 ppm (see Figure 1). In addition, the deviations for the carbon atom C 161.7 ppm are equal to ca. 40 ppm. This data clearly confirms that structure 1 is incorrect, and this conclusion is not surprising.
The authors of [1] comment that the structure proposed for janthinolide A (1) includes a zwitterionic N-peroxide function. This functional group has no precedent in the literature of either natural products chemistry or organic synthesis. In addition, intuitively, this functional group would be expected to be a powerful oxidizing agent, or to fragment to liberate oxygen, and as such would be highly unlikely to be present in a natural product and to have survived the isolation and purification steps employed. The authors presented no direct evidence for this functional group.
The molecular formula C23H32N2O7 and the NMR data (Table 1) originally reported [1] were entered in to Structure Elucidator.
Table 1. NMR spectroscopic data of janthinolide A.
| Label | δC | XHn | δH | H to C HMBC |
| C 2 | 161.7 | C | ||
| C 3 | 122.2 | C | ||
| C 4 | 152 | C | ||
| C 5 | 166 | C | ||
| C 7 | 132.1 | CH | 7.34 | |
| C 8 | 126.1 | C | ||
| C 9 | 159.6 | C | ||
| C 10 | 115.1 | CH | 6.89 | |
| C 12 | 131.5 | CH | 7.34 | |
| C 15 | 64.8 | CH2 | 4.64 | |
| C 16 | 120.2 | CH | 5.5 | |
| C 17 | 141 | C | ||
| C 18 | 16.424 | CH3 | 1.724 | |
| C 19 | 36.2 | CH2 | 2.33 | |
| C 19 | 36.2 | CH2 | 2.13 | |
| C 20 | 29.2 | CH2 | 1.65 | |
| C 20 | 29.2 | CH2 | 1.48 | |
| C 21 | 77.8 | CH | 3.32 | |
| C 22 | 73.3 | C | ||
| C 24 | 23.1 | CH3 | 1.18 | |
| C 25 | 26.3 | CH3 | 1.19 | |
| C 30 | 52.6 | CH3 | 3.86 | C 5 |
| C 31 | 9.1 | CH3 | 2.13 | C 4, C 2 |
| OH | 2.714 | |||
| OH | 3.185 | |||
| OH | 8.77 | C 4, C 2 | ||
| NH | 8.29 | C 3, C 4, C 2, C 5 |
A molecular connectivity diagram (MCD) was created by the program and also the ”green fragment” shown in Figure 1 was defined to be included in all structures (see Figure 2).
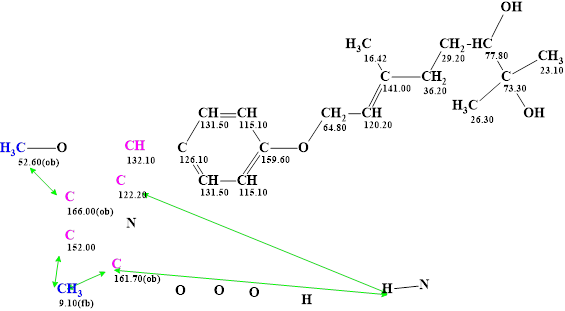
Figure 2. Molecular connectivity diagram (MCD) of janthinolide A. The MCD contains the fragment of the janthinolides A structure colored in green (Figure 1). Hybridizations of carbon atoms are marked by the corresponding colors: sp2 – violet, sp3 – blue. Labels “ob” and “fb” are set by the program to carbon atoms for which neighboring with a heteroatom is either obligatory (ob) or forbidden (fb). Green arrows show the HMBC connectivities.
Strict structure generation combined with 13C chemical shift prediction and structure filtering was carried out. Experimental chemical shifts were automatically assigned to carbon atoms of structures generated. As a result, 55 structures were obtained in 12 s. The six top structures of the output file ranked in increasing order of average deviations are presented in Figure 3.
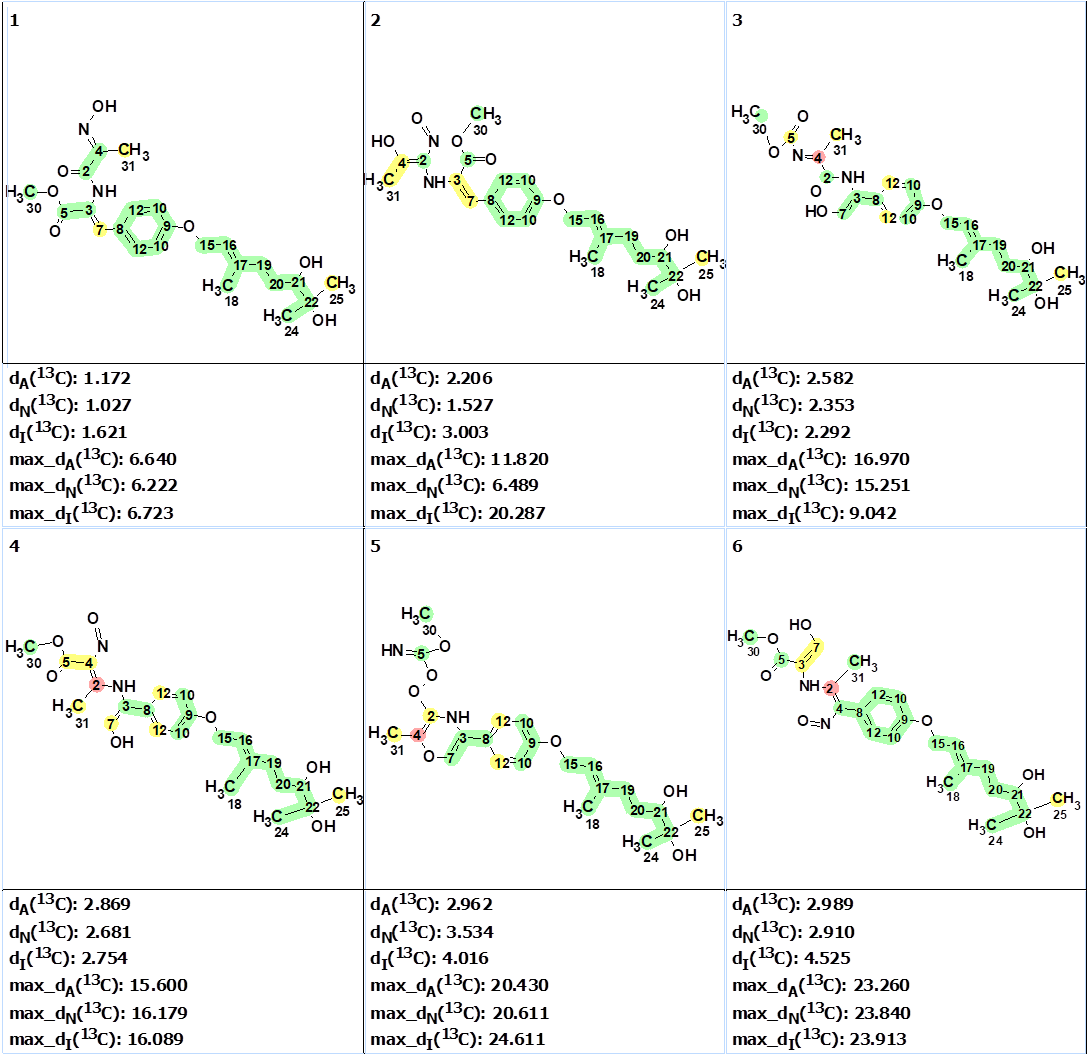
Figure 3. The six top ranked structures of the output file. 13C chemical shift prediction was carried out using the HOSE code-based method, the neural networks, and the incremental approach. Average deviations of 13C chemical shifts determined by these methods are denoted as dA, dN and dI correspondingly. The structures are ranked in increasing order of dA(13C ) values. Each atom is colored to mark a difference between its experimental and calculated 13C chemical shifts. The green color represents a difference between 0 to 3 ppm, yellow was >3 to 15 ppm, red > 15 ppm.
The two best structures are shown in Figure 2.
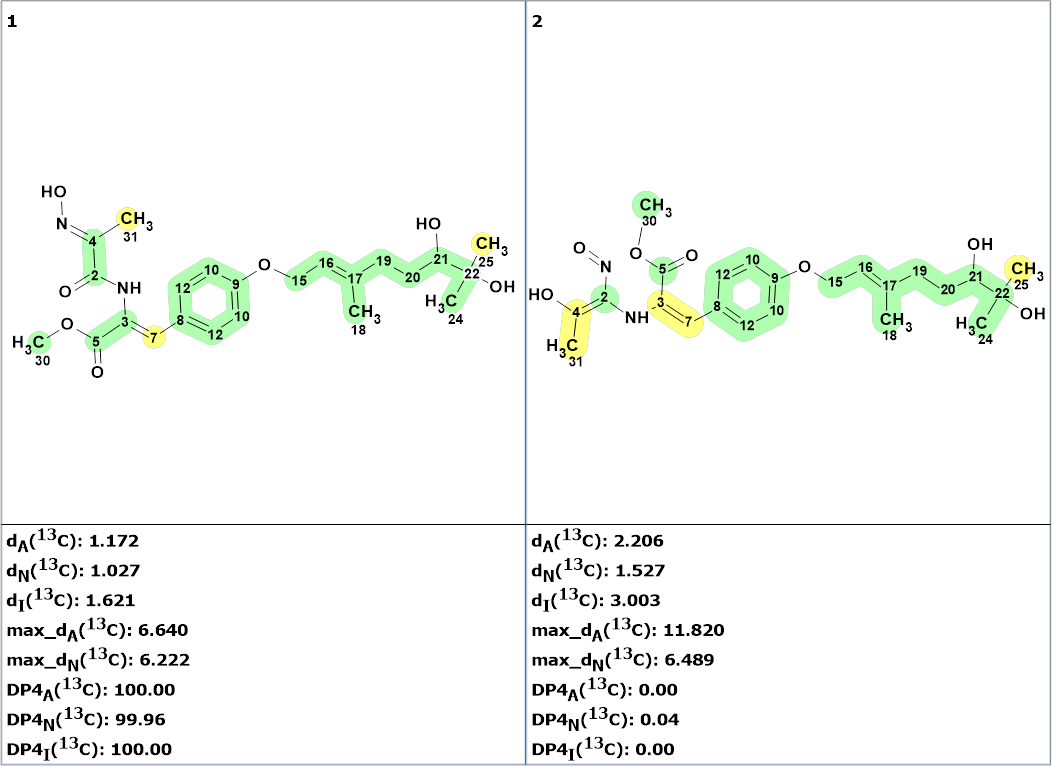
Figure 4. The two top structures of the ranked output file. 13C chemical shift prediction was carried out using the HOSE code-based method, the neural networks, and the incremental approach. Average deviations of 13C chemical shifts determined by these methods for each structure are correspondingly denoted as follows: dA (HOSE code-based), dN (neural networks) and dI (incremental approach). Values DP4A(13C ), DP4N(13C ), and DP4I(13C ) are the DP4 derived probabilities that a given structure is correct.
The first structure is an a-oximocarbonyl compound. Such compounds are well known in the natural products literature. The calculated DP4 probability for this structure was equal to ~100%. Its validity was confirmed by DFT DU8ML calculations. The second structure must be rejected not only because of the larger average deviations, but also from chemical considerations: it is a vinyl nitroso compound which would be unusual for a natural product. In addition, it would appear to be an unstable tautomer.
The authors of [2] revealed the following interesting coincidence: Janthinolide C, an analog of Janthinolide A, was isolated from P. griseofulvum, in which the diketopiperazine-derived moiety has seemingly undergone both oxidation and ring opening to give an a-oximocarbonyl moiety [3]. Janthinolide C also possesses the same molecular formula as janthinolide A. Authors [2] concluded that janthinolide C had the same structure as the revised structure of janthinolide A . Comparison of the reported 13C NMR data of janthinolide A (1) and of janthinolide C, as well as the calculated 13C NMR data for janthinolide A showed a very high degree of similarity. Comparison of the reported 1H NMR data for janthinolides A and C also showed strong similarity.
The revised structure of janthinolide A with the assigned 13C chemical shifts is shown below:
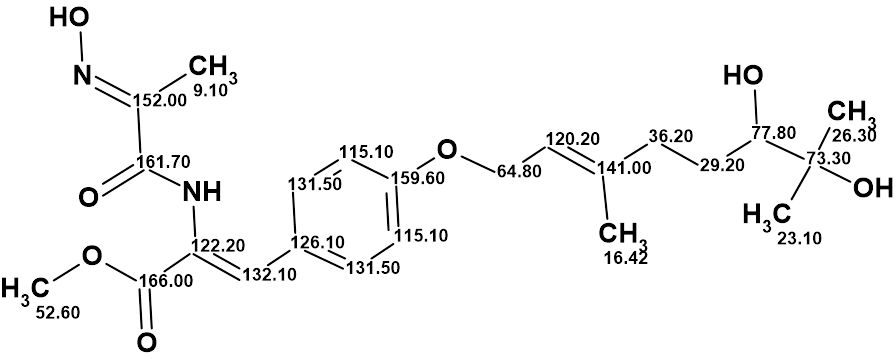
Thus, the correct structure of janthinolide A was determined and in addition it was shown that structure of janthinolide C suggested in [3] was identical to janthinolide A.
References
- C. Xue, T. Li, Z. Deng, H. Fu, W. Lin. (2006). Janthinolide A-B, two new 2,5-piperazinedione derivatives from the endophytic Penicillium janthinellum isolated from the soft coral Dendronephthya sp. Chunmei. Pharmazie., 61, 1041–1044.
- R.W. Bates, M. Elyashberg, A. G. Kutateladze, C. M. Williams. (2023). Reassignment of the Structure of Janthinolide A. Eur. J. Org. Chem., 23, e202300910. doi.org/10.1002/ejoc.202300910.
- Y. Zang, Y. Gong, X. Chen, H. Wen, C. Qi, C. Chen, J. Liu, Z. Luo, J. Wang, H. Zhu, Y. Zhang. (2021). Piperazine-2,5-dione derivatives and an α-pyrone polyketide from Penicillium griseofulvum and their immunosuppression activity. Phytochemistry, 186, 112708.


