February 2, 2024
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
(±)-Salvicatone A
The Salvia genus, a prominent member of the Labiatae family, encompasses more than 1000 species distributed across temperate and warmer regions worldwide. Some of these species have been utilized in traditional medicine for millennia. By 2011, researchers had isolated 773 compounds from 134 Salvia species. In the last decade, ongoing research on the Salvia genus has unveiled an increasing number of uniquely structured compounds.
In order to explore the novel bioactive constituents of this species, Wang et al [1] conducted a systematic phytochemical investigation on the roots and rhizomes of SCFT. As a result, they isolated a rare pair of C27-meroterpenoid enantiomers, (±)-salvicatone A (1), featuring a unique 6/6/6/6/6-pentacyclic carbon skeleton.
1
The authors identified the structure of this compound in two stages. In the first step, they used 1H, 13C , HMBC and COSY spectra and a molecular formula to deduce the structure of the unknown compound using the traditional methods. In the second step, the authors used ACD/Structure Elucidator Suite to validate the proposed structure. Structure Elucidator created the molecular connectivity diagram (MCD), assuming that the molecule may contain a maximum bond multiplicity equal to 3. The edited MCD is shown in Figure 1. Checking the MCD for the presence of nonstandard correlations (NSCs) showed that all HMBC and COSY correlations are of standard length (nJCH,HH, n=2-3). Then all atom hybridizations were set as in the proposed structure 1, and in spite of the results of MCD analysis, 13 potential NSCs were drawn manually in the MCD. No explanation of this decision leading to increasing the time of structure generation was given in the article. Two benzene rings and a carbonyl group were artificially defined (drawn manually in MCD) to reduce the amount of computations in the program.
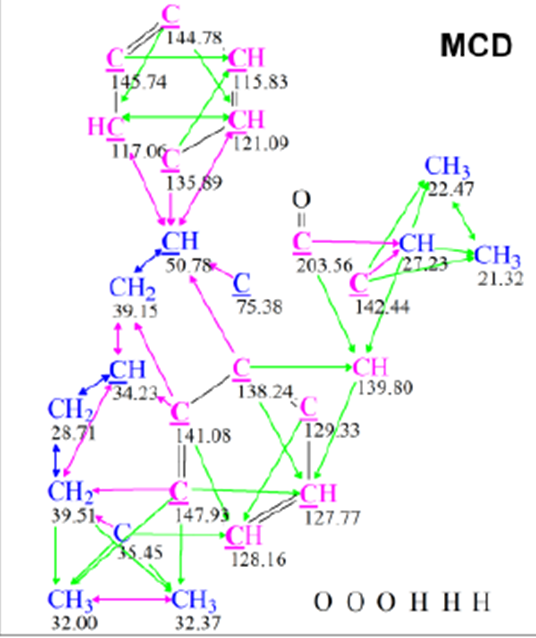
Figure 1. Molecular connectivity diagram. The states of atom hybridization are designated in its own specific color as sp3–blue, sp2–violet. Standard HMBC and COSY correlations are marked with green (HMBC) and blue (COSY) arrows. Nonstandard correlations are marked with violet arrows.
As a result of Strict Structure Generation, 5591 structures were generated in 21 s, and 52 structures were saved after spectral filtering. The prediction of 13C chemical shifts was carried out for each candidate using the three empirical methods provided by Structure Elucidator (HOSE, dA; incremental approach, dI; and neural networks, dN), and then the output file was ranked in increasing order of the average deviation (dA) of 13C chemical shifts between the experimental and calculated values. The eight top ranked structures are shown in Figure 2.
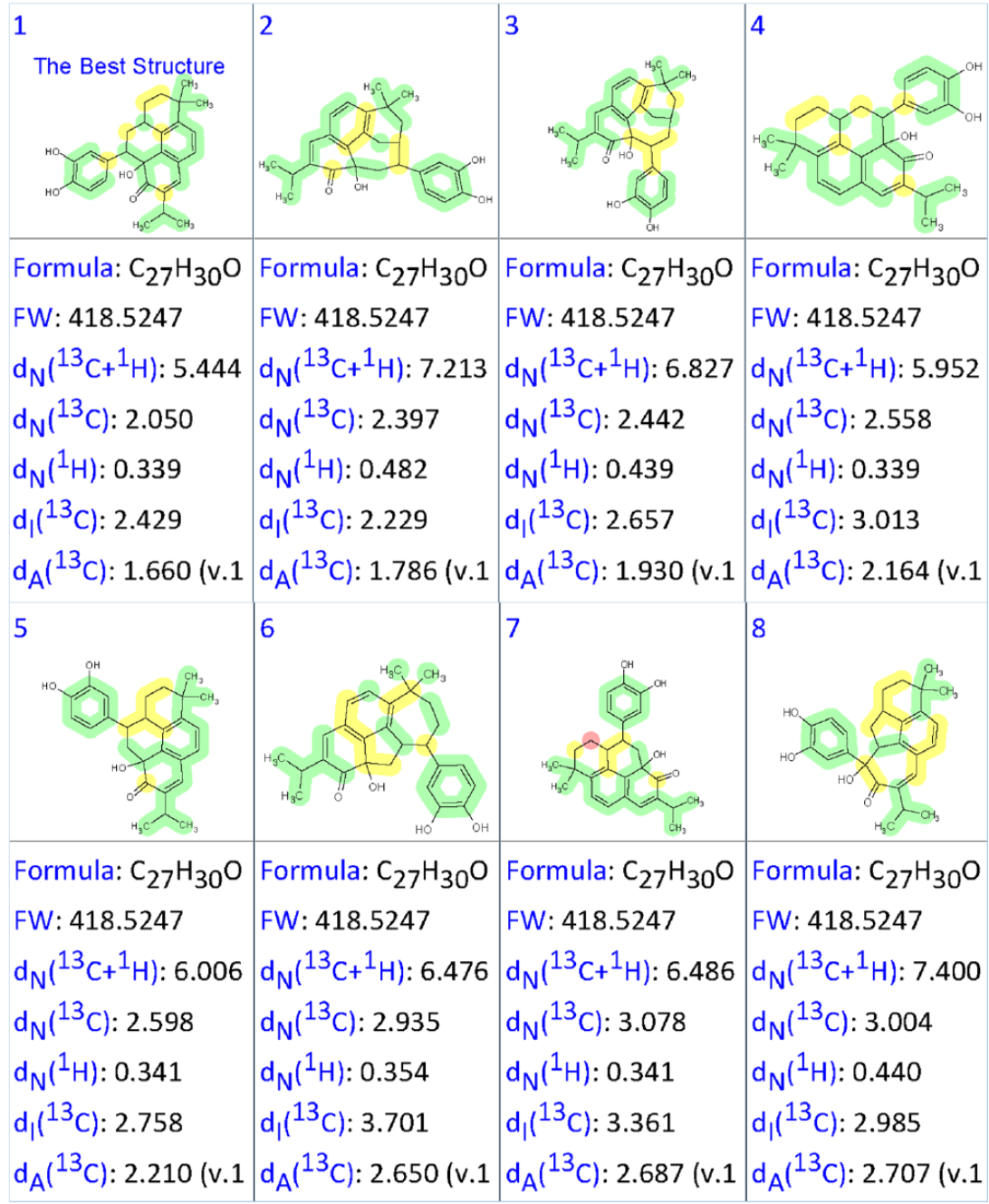
Figure 2. The eight top ranked structures of the output file presented into Supporting Information of work [1]
We see that the best structure is identical to that deduced by authors [1] as a result of manual analysis of combined HMBC and COSY data. Using DFT based calculations, the authors determined the most probable conformer for which the average deviation (MAE) was found to be 1.31 ppm. This MAE value is typical for correct structures, which confirmed the CASE answer.
ACD/Structure Elucidator provides different strategies for NMR based structure elucidation. The researcher has the possibility to select any strategy which is more attractive to them. However, a standard strategy tested on many hundreds of solved problems and described in textbook [2] looks optimal for the overwhelming majority of cases. More than a hundred examples solved with this strategy can be found at the ACD/Labs web site. Therefore, we repeated the solution described in [1] applying the recommended approach.
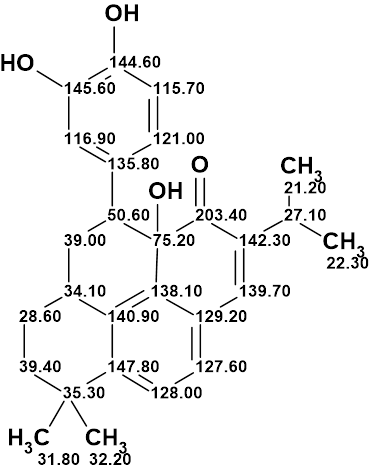
The 1D NMR spectra and key 2D NMR correlations (Table 1) presented in the article, accompanied with the molecular formula C27H30O4, were entered into Structure Elucidator and the molecular connectivity diagram was automatically created (Figure 3).
The IR spectrum of compound 1 (Figure 4) shows that no absorption is observed in the area between 2000 and 2500 cm-1, which allows one to suppose that the unknown compound does not contains a triple bond. Therefore, the maximum possible bond multiplicity was set equal to 2 in the options for MCD creation.
Table 1. NMR spectroscopic data of compound 1
| C/X Label | δC | δCcalc (HOSE) | XHn | δH | COSY | H to C HMBC |
| C 1 | 34.1 | 36.1 | CH | 3.47 | 1.26, 1.49 | C 10 |
| C 2 | 28.6 | 24.52 | CH2 | 1.49 | 1.76, 3.47 | |
| C 2 | 28.6 | 24.52 | CH2 | 2.02 | ||
| C 3 | 39.4 | 35.51 | CH2 | 1.76 | 1.49 | C 5 |
| C 3 | 39.4 | 35.51 | CH2 | 1.81 | ||
| C 4 | 35.3 | 35.7 | C | |||
| C 5 | 147.8 | 147.84 | C | |||
| C 6 | 128 | 125.65 | CH | 7.48 | 7.19 | C 4, C 8, C 10 |
| C 7 | 127.6 | 128.33 | CH | 7.19 | 7.48 | C 9, C 14, C 5 |
| C 8 | 129.2 | 131.39 | C | |||
| C 9 | 138.1 | 137.3 | C | |||
| C 10 | 140.9 | 136.05 | C | |||
| C 11 | 75.2 | 75.66 | C | |||
| C 12 | 203.4 | 202.22 | C | |||
| C 13 | 142.3 | 138.39 | C | |||
| C 14 | 139.7 | 139.5 | CH | 7.03 | C 15, C 9, C 12 | |
| C 15 | 27.1 | 27.85 | CH | 2.74 | 0.66, 1.02 | |
| C 16 | 22.3 | 21.83 | CH3 | 0.66 | 2.74 | C 13 |
| C 17 | 21.2 | 21.83 | CH3 | 1.02 | 2.74 | C 13 |
| C 18 | 32.2 | 32.32 | CH3 | 1.37 | C 4 | |
| C 19 | 31.8 | 30.32 | CH3 | 1.38 | C 4, C 3, C 5 | |
| C 20 | 39 | 34.64 | CH2 | 1.26 | 3.47, 3.62 | C 10 |
| C 20 | 39 | 34.64 | CH2 | 2.77 | ||
| C 21 | 50.6 | 48.95 | CH | 3.62 | 1.26 | C 11, C 22, C 9 |
| C 22 | 135.8 | 131.22 | C | |||
| C 23 | 116.9 | 115.7 | CH | 6.03 | C 21, C 25 | |
| C 24 | 145.6 | 145.57 | C | |||
| C 25 | 144.6 | 144.13 | C | |||
| C 26 | 115.7 | 115.86 | CH | 6.35 | 5.83 | C 22, C 24 |
| C 27 | 121 | 120.37 | CH | 5.83 | 6.35 | C 23, C 25 |
| O 1 | OH | 5.56 | C 11, C 9 | |||
| O 2 | OH | 8.45 | C 24 | |||
| O 3 | OH | 8.39 | C 26, C 25 |
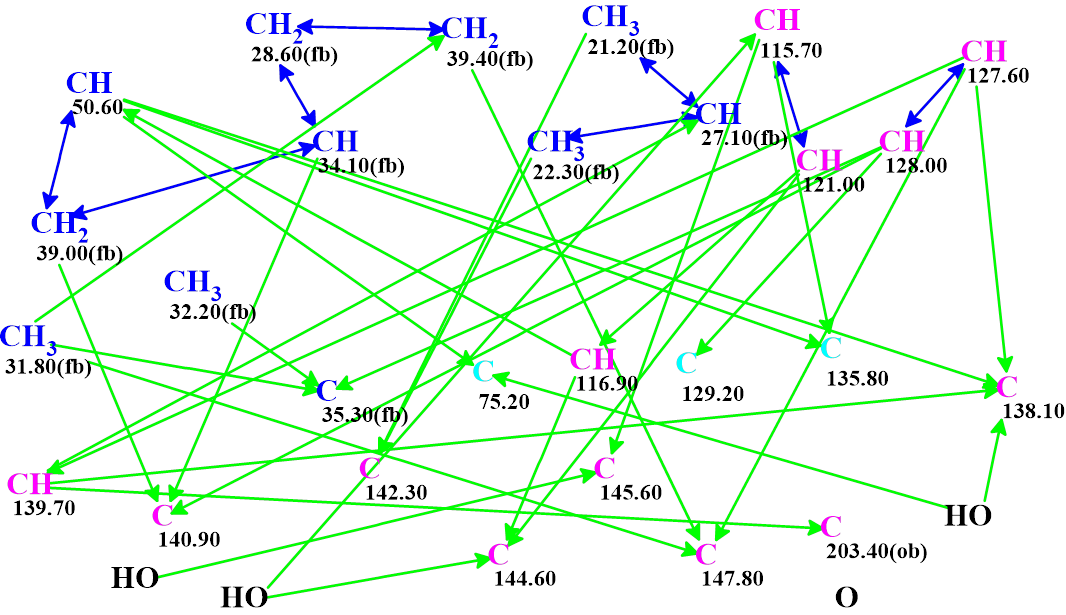
Figure 3. MCD automatically created from the spectroscopic data shown in Table 1. Hybridizations of carbon atoms are marked by corresponding colors: sp2 – violet, sp3 – blue, not sp – light blue. Labels “ob” and “fb” are set by the program to carbon atoms for which neighboring with heteroatom is either obligatory (ob) or forbidden (fb). HMBC connectivities are marked by green arrows, while COSY connectivities – by blue arrows.
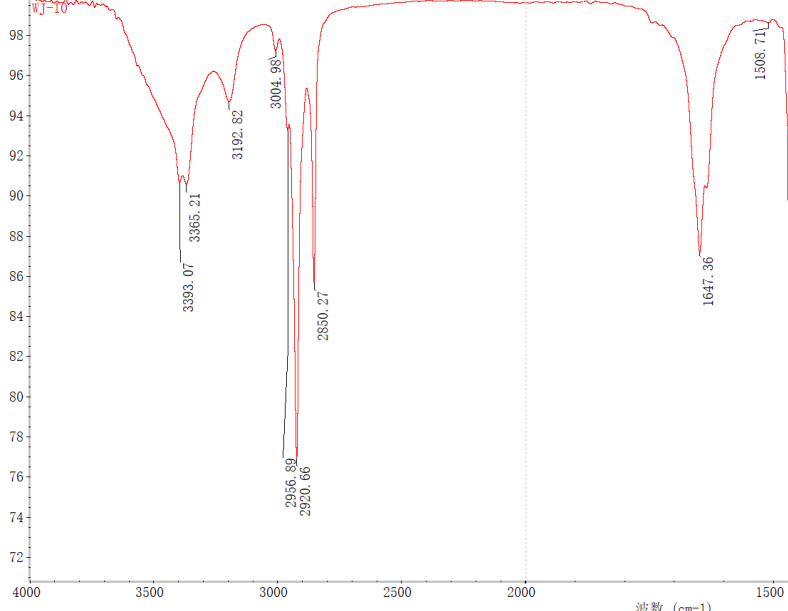
Figure 4. A fragment of the IR spectrum of compound 1 recorded in MeOH.
No edits manual were made on the MCD, and strict structure generation was initiated which was completed with the following results: k=25 → (Filter) → 7 → (Duplicate Removal) → 7, tg = 3 s. 13C chemical shift predictions were performed as described previously and the structures were ranked in increasing order of the average deviation (dA). The ranked output file is presented in Figure 5.
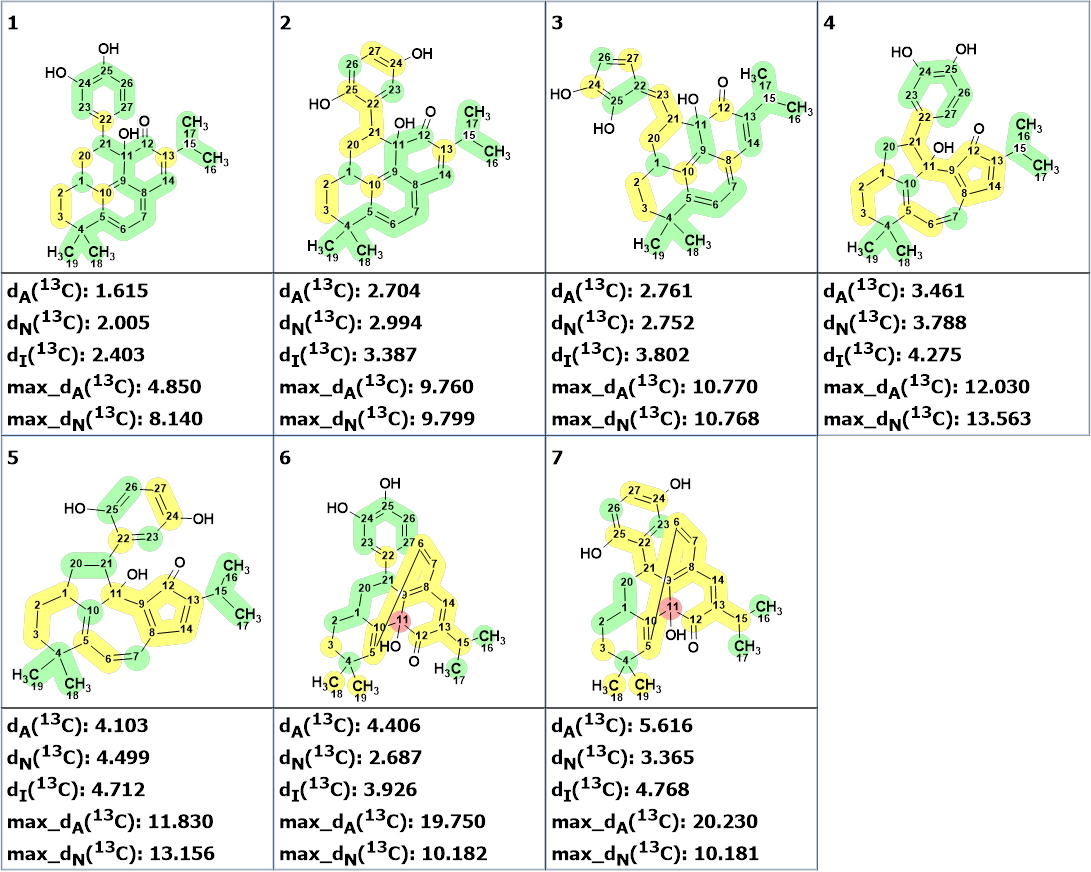
Figure 5. The ranked output file. Red color indicates atoms for which the deviation between experimental and calculated 13C chemical shifts exceeds 15 ppm.
We see that the top ranked structure coincides with structure of salvicatone A determined in article [1]. Figure 6 shows the three first ranked structures with calculated empirical DP4 probabilities.
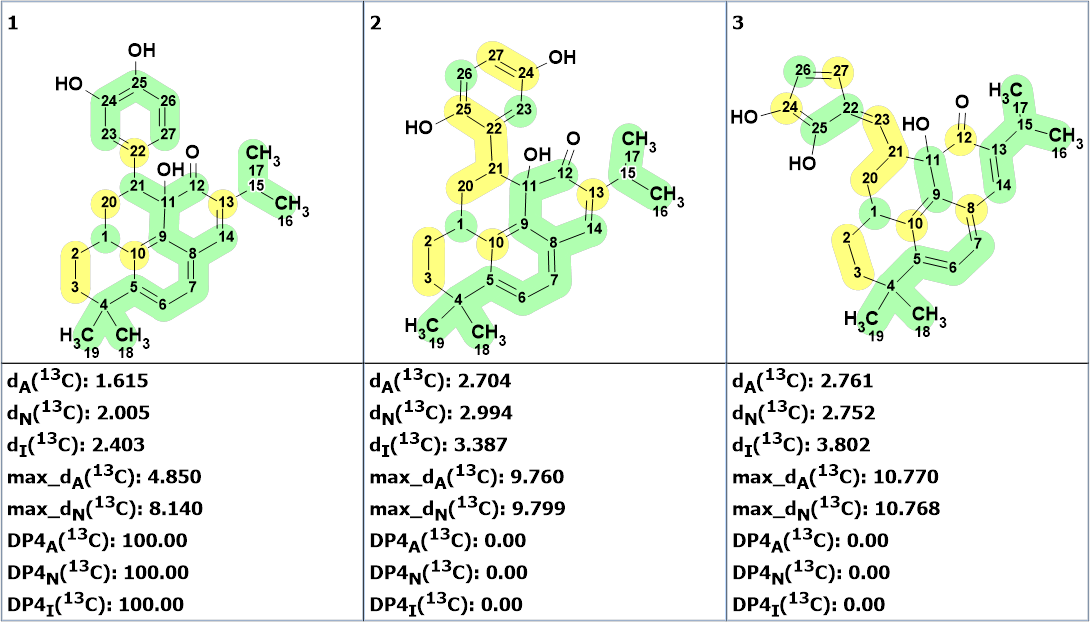
Figure 6. Three top ranked structures with calculated empirical DP4 probabilities.
Figure 6 shows that the probability of the structure assigned as salvicatone A is equal to 100%. For comparison with the solution found in [1], we decided to see which solution would be obtained if the presence of triple bond was allowed. The MCD automatically created for this case by the program is shown in Figure 7.
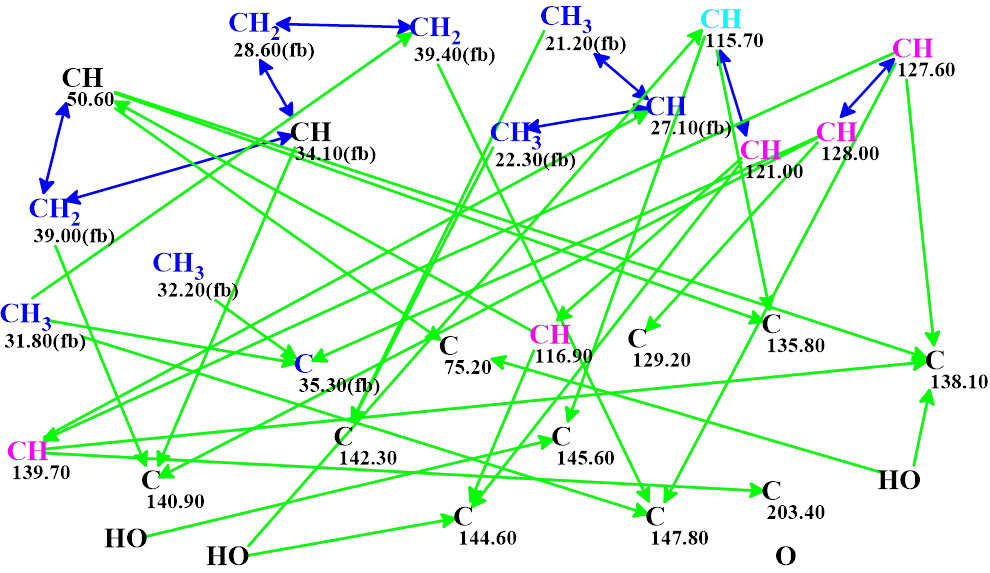
Figure 7. The automatically created MCD, assuming that a triple bond may exist in the molecule. Carbon atoms whose hybridization is undefined are marked by black color.
As can be expected, the structure generation time was longer than in previous run: it took 1 min 6 s, but the final output file was the same as presented in Figure 5.
In this example we showed that the structure of a complex natural product, salvicatone A, containing a unique 6/6/6/6/6-pentacyclic carbon skeleton was unambiguously elucidated fully automatically (without any user intervention) in several seconds. The structure of this compound with assigned 13C chemical shifts is shown below.

References
- Wang, D.-D.; Zhang, R.; Tang, L.-Y.; Long, G.-Q.; Yan, H.; Yang, Y.-C.; Guo, Z.-F.; Zheng, Y.-Y.; Wang, Y.; Jia, J.-M.; Wang, A.-H. (2023). (±)-Salvicatone A: A Pair of C27-Meroterpenoid Enantiomers with Skeletons from the Roots and Rhizomes of Salvia castanea Diels f. tomentosa Stib. J. Org. Chem. https://doi.org/10.1021/acs.joc.3c01664
- M.E. Elyashberg, A.J. Williams. Computer-based Structure Elucidation from Spectral Data. The Art of Solving Problems. Springer, Heidelberg, 2015, 454 p. http://www.springer.com/978-3-662-46401-4


