July 1, 2015
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
A Heterodimer from P. kaurabassana
The genus Pyrenacantha (Icacinaceae) is composed of about 30 different species, of which only three are found in southern Africa: P. grandif lora Baill., P. scandens Planch, and P. kaurabassana Baill. Pyrenacantha species are used widely to treat infectious diseases such as ulcers, diarrhea, herpes, and AIDS in local traditional medicine.
Of these, P. kaurabassana is a dioecious liana. Few constituents have been isolated from P. kaurabassana, and to date only two xanthones (1* and 2*, below) with weak anti-HIV activity and a chrysene derivative have been described, by Omolo and co-workers [1].
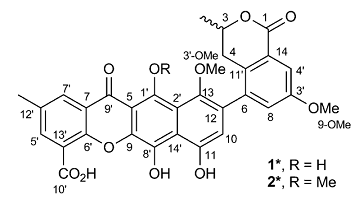
Boudesocque-Delaye et al [2] reported a phytochemical study of the tubers of P. kaurabassana, with the structures of two polyketide heterodimers (1 and 2, below) determined based on spectroscopic data (including a 2D INADEQUATE spectrum) interpretation, along with an evaluation of their antibacterial activity. On the basis of the data obtained, the structures 1* and 2* that were previously proposed for compounds 1 and 2 were revised.
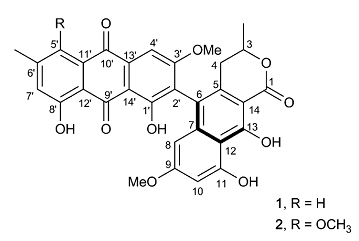
In the second study [2], two heterodimers comprising an anthraquinone moiety linked to a 3-methylbenzodihydroisocoumarin unit were isolated (125 mg of compound 1 and 55 mg of compound 2) from P. kaurabassana tubers (2 g of CH2Cl2 extract). The NMR spectroscopic data presented therein were used by us to challenge the ACD/Structure Elucidator system, by attempting to successfully elucidate structure 1.
Compound 1 gave the molecular formula C31H24O10 on the basis of the 13C NMR data and an HRESIMS ion m/z 555.1298 [M – H]– (calculated 555.1291). The UV spectrum showed maximum absorption bands at 224, 264, 308, and 372 at nm, suggesting the presence of a quinone functionality. In view of the large number of nonprotonated aromatic carbons, the authors [2] came to the conclusion that the structure of compound 1 could not be established using 2D 1H –13C NMR chemical shift correlations. Indeed, the ratio n(skeletal atoms)/n(protons) is equal to 1.7. According to “Crews rule”, if the ratio is close to 2, it may be difficult and frequently impossible to unequivocally elucidate a structure based solely on HMBC and COSY NMR data and molecular formula information. Thus, a 2D INADEQUATE experiment at natural 13C abundance was performed on compound 1 as 13C –13C NMR chemical shift correlations could provide reliable structural data here.
The NMR spectroscopic data adopted from Boudesocque-Delaye et al [2] are presented in Table 1. The atom numbering set by ACD/Structure Elucidator are displayed on structure 1.
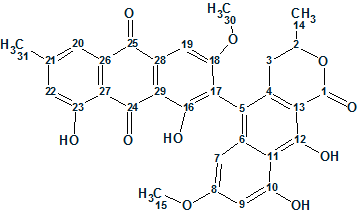
1
Table 1. Heterodimer 1. HMBC and INADEQUATE NMR spectroscopic data.
n(H) indicates the number of protons attached to carbon atoms existing in the first sphere around a given carbon. This value is determined from multiplicities and coupling constants measured in 1H NMR.
| Label | δC | δC calc | XHn | δH | n(H) | C HMBC | INADEQUATE |
| C 1 | 170.9 | 170.03 | C | C 13 | |||
| C 2 | 75.6 | 75.21 | CH | 4.69 | u | C 4 | C 14, C 3 |
| C 3 | 32.2 | 30.53 | CH2 | 2.75 | 1 | C 14, C 5, C 13, C 2, C 4 | C 2, C 4 |
| C 3 | 32.2 | 30.53 | CH2 | 2.63 | u | C 2, C 4 | |
| C 4 | 133.1 | 139.64 | C | C 3, C 13, C 5 | |||
| C 5 | 116.8 | 116.6 | C | C 17, C 4, C 6 | |||
| C 6 | 139 | 140.99 | C | C 7, C 11, C 5 | |||
| C 7 | 98.2 | 96.76 | CH | 6.19 | 0 | C 9, C 5, C 8, C 11 | C 6, C 8 |
| C 8 | 162.3 | 163.18 | C | C 7, C 9 | |||
| C 9 | 100.2 | 101.03 | CH | 6.54 | 0 | C 7, C 11, C 8, C 10 | C 10, C 8 |
| C 10 | 158.9 | 160.72 | C | C 9, C 11 | |||
| C 11 | 108 | 108.14 | C | C 6, C 10, C 12 | |||
| C 12 | 162.6 | 163.17 | C | C 13, C 11 | |||
| C 13 | 99.1 | 102.12 | C | C 4, C 12, C 1 | |||
| C 14 | 19.9 | 20.81 | CH3 | 1.35 | 1 | C 3, C 2 | C 2 |
| C 15 | 55 | 55.28 | CH3 | 3.66 | 0 | C 8 | |
| C 16 | 161.2 | 161.96 | C | C 29, C 17 | |||
| C 17 | 118.9 | 118.18 | C | C 5, C 16, C 18 | |||
| C 18 | 164.2 | 164.06 | C | C 19, C 17 | |||
| C 19 | 103.2 | 103.22 | CH | 7.51 | 0 | C 16, C 5, C 18, C 29, C 17, C 28, C 25 | C 28, C 18 |
| C 20 | 120.7 | 121.5 | CH | 7.52 | 0 | C 22, C 31, C 24, C 25, C 21, C 27, C 23 | C 26, C 21 |
| C 21 | 148.7 | 148.85 | C | C 31, C 20, C 22 | |||
| C 22 | 124.1 | 124.8 | CH | 7.12 | 0 | C 26, C 23, C 20, C 31, C 27, C 24 | C 21, C 23 |
| C 23 | 161.6 | 162.7 | C | C 27, C 22 | |||
| C 24 | 190.6 | 191.25 | C | C 29, C 27 | |||
| C 25 | 181.1 | 182.05 | C | C 26, C 28 | |||
| C 26 | 132.6 | 133 | C | C 27, C 20, C 25 | |||
| C 27 | 113.3 | 113.6 | C | C 26, C 23, C 24 | |||
| C 28 | 134.8 | 135.2 | C | C 19, C 29, C 25 | |||
| C 29 | 110.8 | 115.37 | C | C 28, C 16, C 24 | |||
| C 30 | 56.4 | 56.75 | CH3 | 3.89 | 0 | C 18 | |
| C 31 | 21.3 | 22.2 | CH3 | 2.62 | 0 | C 20, C 21, C 22 | C 21 |
| O 1 | 100* | OH | 12.24 | 0 | C 29, C 16, C 17 | ||
| O 2 | 150* | OH | 11.8 | 0 | C 22, C 27, C 23 |
*Fictitious 17O chemical shifts introduced to define two OH groups which were identified in 1H NMR [2].
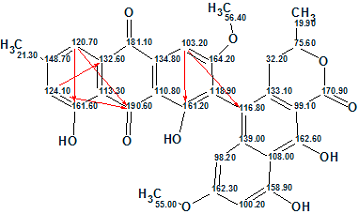
A Molecular Connectivity Diagram (MCD) was created by the software from the data presented in Table 1:
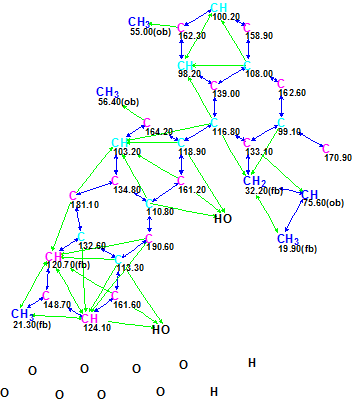
Figure 1. An initial Molecular Connectivity Diagram for Heterodimer 1.
MCD overview. The MCD shows that the INADEQUATE data the main part of the molecular skeleton (blue arrows) to be elucidated. It is also easy to visually detect that the HMBC spectrum contains at least four nonstandard 4JCH-type correlations. 10 carbon atoms are colored in light blue color, which means that their hybridization was not determined exactly by the program (sp3 or sp2, but not sp). For some carbon atoms, the possibility of neighboring with a heteroatom was automatically set from 13C and 1H chemical shift analysis (ob – obligatory, fb – forbidden).
Checking the MCD for the presence of contradictions (which took 7s) showed that the HMBC data contain nJCH, n>3 correlations (as expected). The data presented in n(H) column were not added to the MCD, and no MCD edits were made. Fuzzy Structure Generation was initiated in the mode “Set Options Automatically”. Generation was followed by 13C chemical shift prediction and structure filtering (structures for which average deviations d>4 ppm were rejected). The results were: k=268 → 1, tg = 0.4 s, 6 from 14 correlations have been extended during generation and only 4 from 3003 possible connectivity combinations were used during generation. The single output structure is shown below (red arrows indicate connectivities of non-standard lengths) which is characterized by 13C average deviations dA=1.11, dN=1.73 and dI=1.70 ppm.
This structure coincides with structure 1, and the automatic chemical shift assignments are the same as those made by Boudesocque-Delaye et al [2]. This result clearly demonstrates the advantage of usage INADEQUATE in combination with HMBC in a CASE system to elucidate a very challenging “Crews’ structure”. We also expected that the recently suggested LR-HSQMBC (Long-Range Heteronuclear Single Quantum Multiple Bond Correlation) experiment (see [3,4] and review [5]) would be also helpful to solve the considered problem using Structure Elucidator.
We decided that it would be interesting to use this example for getting answers to the following two questions:
- Would it be possible to correctly elucidate structure 1 using only HMBC data presented in Table 1 as input to Structure Elucidator Suite?
- Would it be possible to correctly elucidate structure 1 using only INADEQUTE data presented in Table 1 as input to Structure Elucidator Suite?
With this in mind, the INADEQUTE spectrum was switched off and a new MCD was created from only the HMBC data. Our experience [6] shows that it is not possible to complete Fuzzy Structure Generation in a manageable time from the molecular formula C31H24O10 (“Crews’ structure” of 41 skeletal atom), HMBC data and an un-edited MCD containing 10 light blue carbon atoms at the presence of 6 nonstandard connectivities. This means that additional information should be introduced. Therefore, the HMBC pattern available from Supporting Information was compared with the HMBC data displayed in Table 1. The correlations whose intensities were small, as well as those which were invisible in the available picture, were marked as 2-4JCH in the software input. In total, eight nonstandard correlations including those six that had been detected by the program earlier were introduced. Then the MCD was again created and edited taking into account well known empirical spectrum-structure correlations (NMR characteristic spectral features). Some evident chemical bonds were drawn by hand and n(H) values were set for corresponding carbon atoms. An edited MCD is presented in Figure 2.
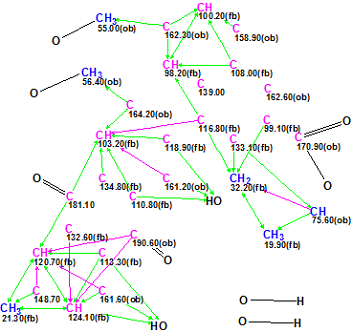
Figure 2. Edited Molecular Connectivity Diagram created from HMBC data only for Heterodimer 1. Nonstandard connectivities are marked by the program by violet color.
No contradictions in HMBC data were detected from MCD checking, and Strict Structure Generation accompanied with 13C chemical shift prediction was initiated. The following results were obtained: k = 46 420 → 9 →6, tg = 48 min. The large number of generated structures and length of structure generation time are explained not only by the presence of 8 2-4JCH correlations, but also by presence of three carbon atoms (C 139.0, C 162.6 and C 170.9) free of any correlations. The output structural file ranked in increasing order of average deviations is shown in Figure 3.
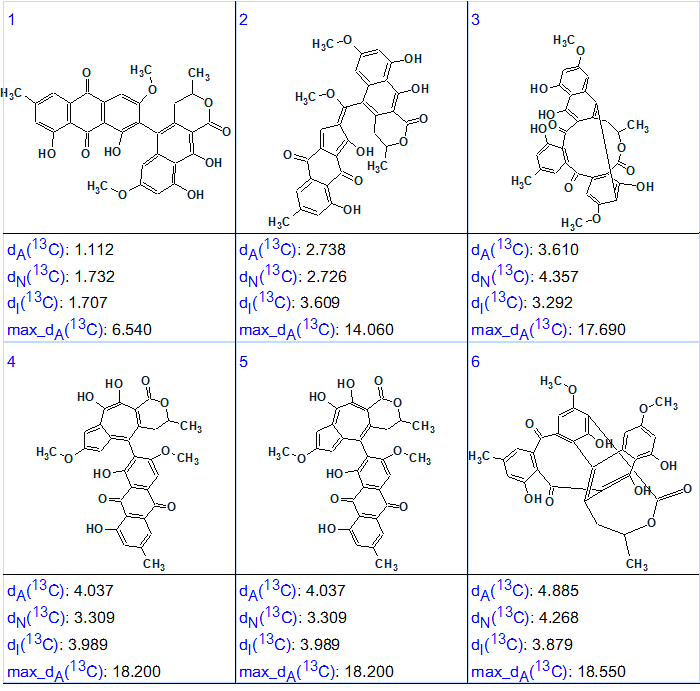
Figure 3. The ranked output structural file generated from HMBC data for Heterodimer 1.
Figure 3 shows that the correct structure was generated and reliably selected by the ranking procedure common for Structure Elucidator Suite. Therefore, as a result of combining both the system’s and spectroscopist’s knowledge (“human-computer symbiosis “) a CASE program was capable of elucidating structure 1 even only from HMBC data in about 50 min (for comparison, INADEQUATE has required for 3 days and 7 hours to be acquired).
To find an answer to the second question posed above, the HMBC spectrum was switched off and an MCD was created from only the INADEQUATE data. As the molecular skeleton was clearly presented in MCD, all light blue carbon atoms were marked as sp2-hybridized, while n(H) values were not used. Structure generation was initiated even without 13C chemical shift prediction. The results were: k = 150 216 → 2292 → 96, tg = 2 m 45 s. The six top ranked structures of the output file are presented in Figure 4.
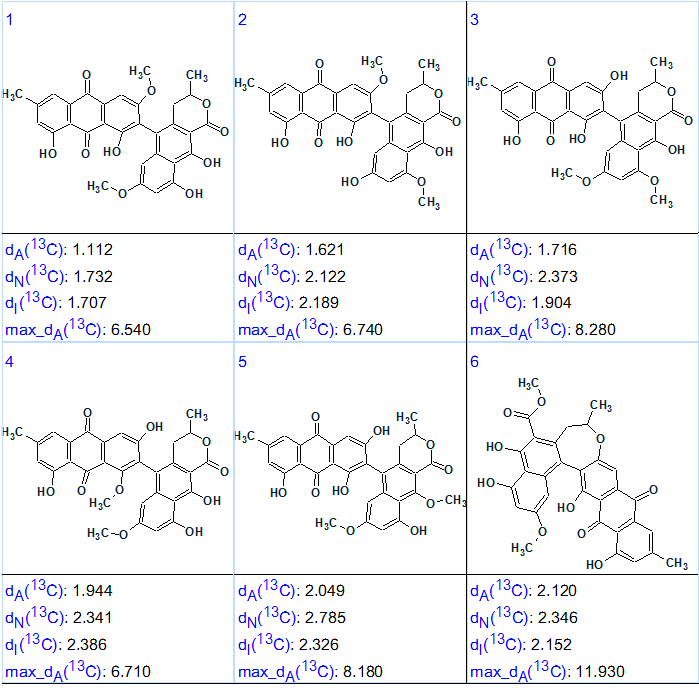
Figure 4. Six top ranked structures of the output file generated from INADEQUATE data for Heterodimer 1.
Figure 4 shows that structure 1 was selected as the best one, but not so reliably as from HMBC data, because many OR substitutions lead to similar structures for which average deviations are of the same order of values. It is interesting to note that when no edits were made to the INADEQUATE-based MCD the resulting structural file was the same (k=96), but a full protocol was another: k = 3 944 492 → 2292 → 96, tg = 1 h 37 m.
This example shows not only the advantages of applying CASE techniques, but also gives interesting insights into different methods of tackling the problem.
References
- J.J. Omolo, V. Maharaj, D. Naidoo, T. Klimkait, H.M. Malebo, S. Mtullu, H.V.M. Lyaruu, C.B. de Koning. (2012). Bioassay-Guided Investigation of the Tanzanian Plant Pyrenacantha kaurabassana for Potential Anti-HIV-Active Compounds. J. Nat. Prod., 75(10):1712–16.
- L. Boudesocque-Delaye, D. Agostinho, C. Bodet, I. Thery-Kone, H. Allouchi, A. Gueiffier, J.M. Nuzillard, C. Enguehard-Gueiffier. (2015). Antibacterial Polyketide Heterodimers from Pyrenacantha kaurabassana Tubers. J. Nat. Prod., 78(4):597–603.
- R.T. Williamson, A.V. Buevich, G.E. Martin, T. Parella. (2014). LR-HSQMBC: A Sensitive NMR Technique To Probe Very Long-Range Heteronuclear Coupling Pathways. J. Org. Chem, 79(9):3887–94.
- K.A. Blinov, A.V. Buevich, R.T. Williamson, G.E. Martin. (2014). The impact of LR-HSQMBC very long-range heteronuclear correlation data on computer assisted structure elucidation. Org. Biomol. Chem., 12:9505–09.
- M. Elyashberg. (2015). Identification and Structure Elucidation by NMR Spectroscopy. Trends in Anal. Chem., 69:88–97.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.


