May 30, 2023
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Saccharobisindole
Actinomycetes are a very well-known group of aerobic and anaerobic Gram-positive mycelial bacteria. They are known to produce a variety of bioactive secondary metabolites. In fact, more than 70% of the currently used antibiotics were originally isolated from Streptomyces, the largest genus of actinomycetes. Rare actinomycetes have also been regarded as potential sources for the discovery of bioactive compounds, including antibiotics. In the past five years, 31% of new bioactive compounds were isolated from rare actinomycete strains, although the Streptomyces genus continues to dominate this field, contributing 65% of the reported bioactive compounds.
The genus Saccharomonospora, a rare actinomycete, was first described in 1971. Analysis of the chemical components from the culture broth of the marine bacterium Saccharomonospora sp. CNQ-490 carried out by Fenical and coworkers [1] has yielded three novel compounds, including saccharobisindole (1). Its chemical structure was elucidated by the interpretation of 1D, 2D NMR (HSQC, HMBC and COSY) , and high-resolution mass spectrometry (HR-MS) data.
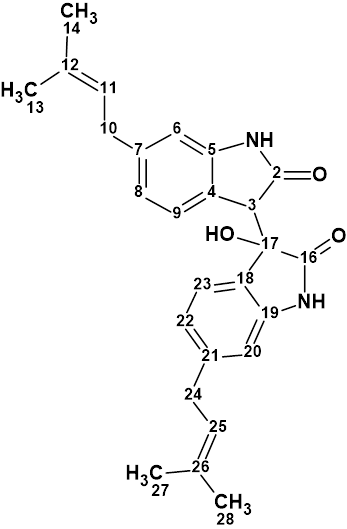
1
Saccharobisindole (1) was obtained as a pale, yellow oil. The molecular formula of this compound (C26H28N2O3) was deduced from high-resolution fast atom bombardment mass spectrometry (HR-FAB-MS) which showed an ion at m/z 417.2181 [M+H]+ (calcd for C26H29N2O3, 417.2178) indicating 14 degrees of unsaturation. The IR spectrum of this compound indicated the presence of a hydroxy group at 3431 cm-1, a carbonyl group at 1676 cm-1, and a double bond at 1638 cm-1.
1D, HSQC and HMBC data tabulated in article [1] along with the molecular formula were entered into ACD/Structure Elucidator (Table 1).
Table 1. NMR spectroscopic data of saccharobisindole (1).
| C/X Label | dC | dCcalc (HOSE) | XHn | dH | M(1H) | H to C HMBC |
| C 2 | 174.6 | 177.76 | C | |||
| C 3 | 51.1 | 54.28 | CH | 3.91 | s | C 17, C 4, C 18, C 5, C 2, C 16 |
| C 4 | 123.2 | 121.84 | C | |||
| C 5 | 143.4 | 144.23 | C | |||
| C 6 | 108.8 | 109.71 | CH | 6.55 | s | C 10, C 8, C 4 |
| C 7 | 141.8 | 142.71 | C | |||
| C 8 | 120.9 | 122.03 | CH | 6.8 | d | C 10, C 6, C 4 |
| C 9 | 126.2 | 126.66 | CH | 7.35 | d | C 7, C 5 |
| C 10 | 33.8 | 33.58 | CH2 | 3.3 | d | C 6, C 8, C 11, C 12, C 7 |
| C 11 | 123.2 | 122.84 | CH | 5.29 | t | C 14, C 13, C 10 |
| C 12 | 131.8 | 133.78 | C | |||
| C 13 | 25.5 | 25.78 | CH3 | 1.72 | s | C 14, C 11, C 12 |
| C 14 | 17.7 | 17.88 | CH3 | 1.69 | s | C 13, C 11, C 12 |
| C 16 | 177.3 | 177.74 | C | |||
| C 17 | 75.4 | 76.19 | C | |||
| C 18 | 125.8 | 128.91 | C | |||
| C 19 | 142.9 | 142.62 | C | |||
| C 20 | 109.4 | 110.18 | CH | 6.53 | s | C 24, C 22, C 18 |
| C 21 | 143.1 | 142.11 | C | |||
| C 22 | 120.8 | 121.48 | CH | 6.45 | d | C 24, C 20, C 18 |
| C 23 | 123.5 | 125.01 | CH | 6.09 | d | C 19, C 21 |
| C 24 | 33.7 | 33.45 | CH2 | 3.19 | d | C 20, C 22, C 25, C 26, C 21 |
| C 25 | 122.9 | 122.41 | CH | 5.21 | t | C 28, C 27, C 24 |
| C 26 | 131.9 | 133.78 | C | |||
| C 27 | 25.4 | 25.78 | CH3 | 1.68 | s | C 28, C 25, C 26 |
| C 28 | 17.6 | 17.88 | CH3 | 1.64 | s | C 27, C 25, C 26 |
| N 1 | NH | 10.08 | s | C 3, C 4, C 5, C 2 | ||
| N 2 | NH | 10.22 | s | C 17, C 18, C 19 | ||
| O 1 | OH | 6.4 | s | C 3, C 17, C 18 |
The Molecular Connectivity Diagram (MCD) created by the program is shown in Figure 1.
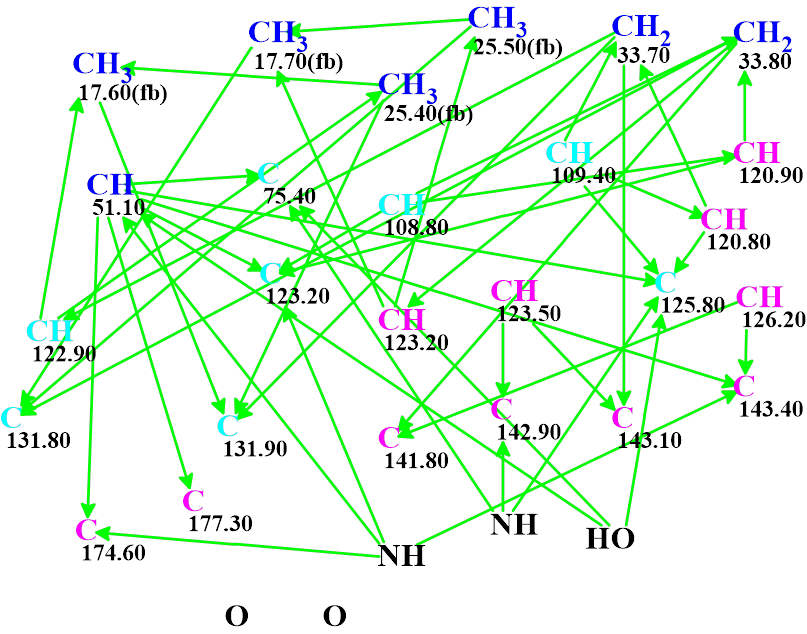
Figure 1. Molecular connectivity diagram. Carbon atoms hybridizations are marked by the corresponding colors: sp2 – violet, sp3 – blue, not sp (sp3 or sp2) – light blue. Labels “fb” are set by the program to carbon atoms for which neighboring with heteroatom is forbidden. The HMBC connectivities are marked by green arrows.
The MCD contains 8 light blue carbon atoms characterized by ambiguous hybridizations. No carbons received the “ob” label, which indicates those atoms which should be connected to a heteroatom. At the same time, as seen in Table 1, reliably determined multiplicities are found for all signals in the proton spectrum. Therefore, the fields “Number of Hydrogens on Neighbor Atoms” were filled in by corresponding figures in the dialog window “Edit Properties of Atom…”. No manual MCD edits were made.
Checking the MCD for consistency showed that there were no contradictions in the 2D NMR data, therefore strict structure generation accompanied with 13C chemical shift prediction and structure filtering was initiated. Results: k = 20 → (structure filtering) → 3, tg = 17 s.
The resulting structural file ranked in increasing order of the average deviations dA(13C ) calculated 13C chemical shifts from the experimental ones is presented in Figure 2.
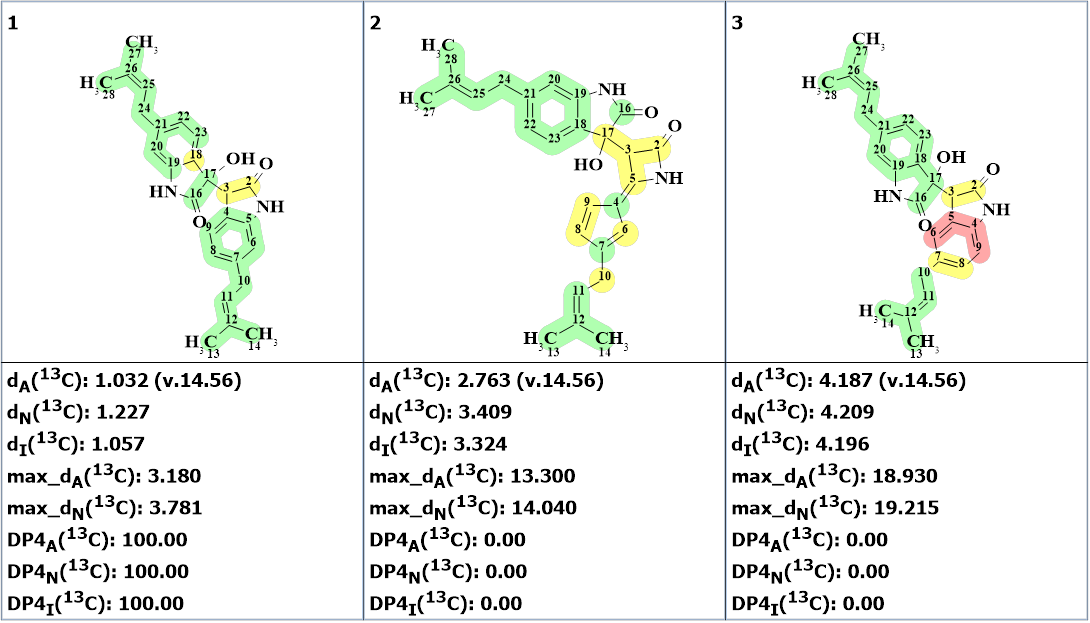
Figure 2. The ranked output file. 13C chemical shift prediction was carried out using the HOSE code-based method, the neural networks, and the incremental approach. Average deviations of 13C chemical shifts determined by these methods are denoted as dA, dN and dI correspondingly. Each atom is colored to mark the difference between its experimental and calculated 13C chemical shifts. The green color represents a difference between 0 to 3 ppm, yellow was >3 to 15 ppm, red > 15 ppm,
We see that the first ranked structure is identical to that suggested by authors[1] and its DP4 probabilities calculated for all three methods of chemical shift prediction are equal to 100%.
It was interesting to see which solution would be obtained if the multiplicities in 1H spectrum were ignored. A new MCD was created, and structure generation was repeated. Results: k = 176 → (structure filtering) → 26 → (duplicate removal) → 7 → (check by Bredt’s rule) → 6, tg = 55 s.
The new ranked output file is presented in Figure 3.
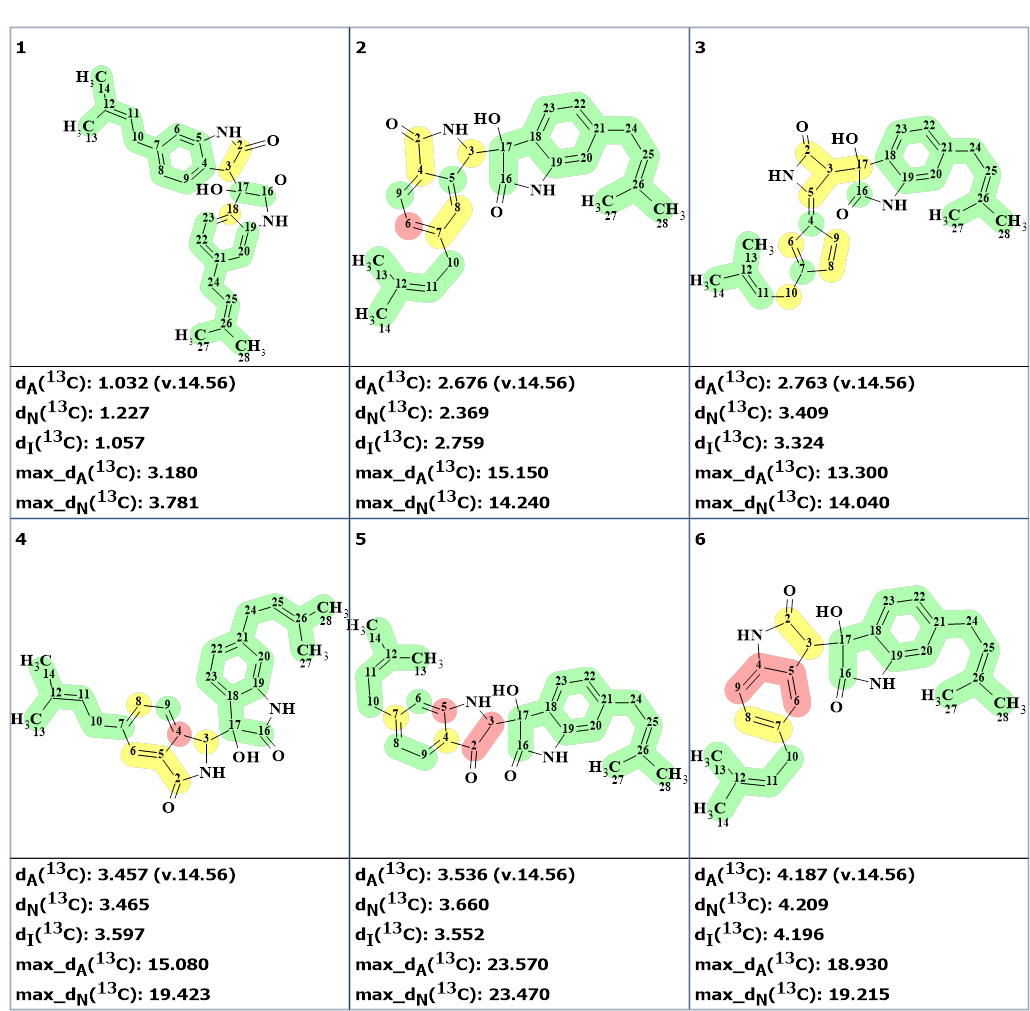
Figure 3. The ranked output file obtained by ignoring multiplicities in 1H NMR spectrum.
Figure 3 shows that the number of generated structures is double while the generation time increased three times. All additional structures contain hydrogen atoms whose multiplicities contradict those indicated in Table 1. The effect would be more noticeable in cases when the expected generation time is longer.
Thus, using ACD/Structure Elucidator allowed us to determine the structure of saccharobisindole in a fully automatic way quickly and reliably. The resulting structure together with the assigned 13C chemical shifts is shown below:

References
- Kim, S.; Le, T. C.; Han, S.-A.; Hillman, P. F.; Hong, A.; Hwang, S.; Du, Y. E.; Kim, H.; Oh, D.-C.; Cha, S.-S.; Lee, J.; Nam, S.-J.; Fenical, W. Saccharobisindole, Neoasterric Methyl Ester, and 7-Chloro-4(1H)-quinolone: Three New Compounds Isolated from the Marine Bacterium Saccharomonospora sp. Marine Drugs 2022, 20, (1), 3. https://doi.org/10.3390/md20010035


