August 1, 2014
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Acremolin
Julianti et al [1] isolated a novel modified base, Acremolin (1), from the culture broth of the marine fungus Acremonium strictum.
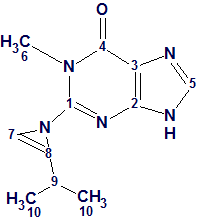
1
Based on the original combined spectroscopic analyses, the structure of this compound was determined to be a methyl guanine base containing an isoprene unit. The presence of a 1H-azirine moiety is unprecedented among natural products, as noted by the authors. [1]
Januar and Molinski [2] were intrigued by this report, as the structure 1 assigned to Acremolin seemed highly surprising to them. They suggested that such an unstable compound would be unlikely to exist in nature, and hypothesized that the acremolin molecule could have one of two possible structures—2 or 3:
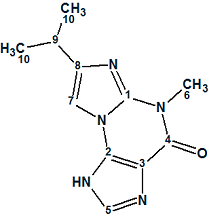 2 |
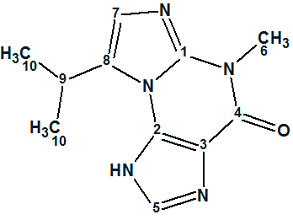 3 |
Independently, Banert [3] proposed the same hypothesis and suggested that structure 2 better fits the NMR data for acremolin, based on analysis of predicted 13C NMR chemical shift increments for2 and 3, but without experimental evidence.
To resolve this issue, Januar and Molinski performed a synthesis of compound 2 and tried (without success) to synthesize compound 3 based on the N2,3-ethenoguanine skeleton. The scheme of five steps synthesis suggested by researchers (Figure 1) is shown below (structure 2 is denoted as 5a and structure 3 as 5b):
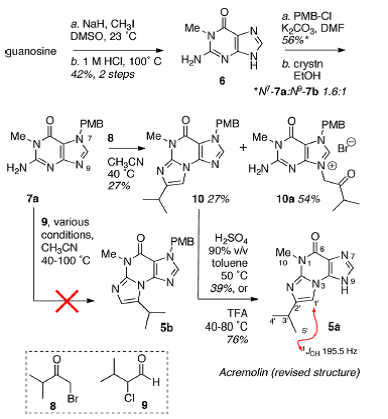
Figure 1. Synthesis performed by Januar and Molinski.
It turned out that the 1H NMR (DMSO-d6, 500 MHz) and 13C NMR spectra of 2 (5a) and other data (UV-VIS, FTIR, HMBC, and HRMS) were identical in every respect with those reported by Julianti and co-workers for acremolin. [1] However compound 5a raises questions of regioisomerism. The location of the isopropyl group in 5a follows from the mechanism of formation of N2,3-ethenoguanine bases. Conceivably, each of the isomers 5a and 5b can be formed.
Doubts regarding the structures of 5a and 5b were removed upon analysis of the experimental 5a 1H–15N HMBC spectrum (Figure 2)—where critical correlations colored in red confirm regioisomer 5a and exclude 5b.
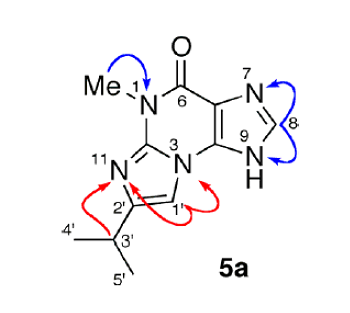
Figure 2. 1H-15N HMBC correlations for structure 5a.
This total synthesis of a hypothetical alternative structure 2 (5a) and its spectroscopic confirmation represents a huge amount of work done by the authors [2] to disprove the original structure 1. As such, we attempted to answer the following question: what solution would be obtained if the original authors [1] used the Computer-Assisted Structure Elucidation (CASE) system ACD/Structure Elucidator Suite for processing the spectroscopic data of the unknown? With this in mind we input 1D NMR spectra and 1H–13C HMBC data of acremolin into the program (Table 1).
Table 1: Spectroscopic NMR data used for acremolin structure elucidation.
| Label | δC | δC calc | XHn | δH | M(J) | C HMBC |
| C 1 | 142.3 | 144.33 | C | |||
| C 2 | 141.6 | 139.48 | C | |||
| C 3 | 108.9 | 111.36 | C | |||
| C 4 | 152.8 | 155.93 | C | |||
| C 5 | 140.5 | 140.79 | CH | 8.16 | u | C 3, C 4, C 2 |
| C 6 | 28.9 | 29.32 | CH3 | 3.57 | u | C 1, C 4 |
| C 7 | 103.2 | 100.04 | CH, NH | 7.38 | u | C 8, C 1 |
| C 8 | 148 | 149.63 | C | |||
| C 9 | 27.7 | 27.81 | CH | 2.88 | u | C 8, C 10, C 7 |
| C 10 | 22.1 | 21.91 | CH3 | 1.25 | u | C 9, C 8 |
From the input data, a Molecular Connectivity Diagram (MCD, Figure 3) was created:
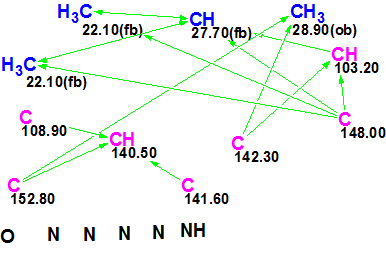
Figure 3. Molecular Connectivity Diagram for acremolin.
No manual edits of the initial, automatically created MCD were made except one: a label “ob” (obligatory) was removed from C 103.2 to avoid unnecessary constraints on structure generation. The software checks the MCD for inconsistencies, but did not detect presence of contradictions in HMBC data, so Strict Structure Generation was initiated. The program produced 81 exotic structures with large average deviations (the best one with d=3–4 ppm) in several seconds. This result hints at the presence of latent nonstandard correlations (NSC). Therefore the next run was performed using Fuzzy Generation Mode with parameters m=1, a=1, i.e., we checked the simplest hypothesis that HMBC data contained at least one NSC of 4JCH type. The result: k = 17004 → 12468 → 498, tg= 1 m 43 s.
Next we performed 13C chemical shift prediction and ranked the output file in ascending order of average deviation values. Both the original structure 1 and the conceivable regioisomer of the revised structure 2 (5a) were generated and saved. Their positions in the ranked structural file are shown in Figure 4.
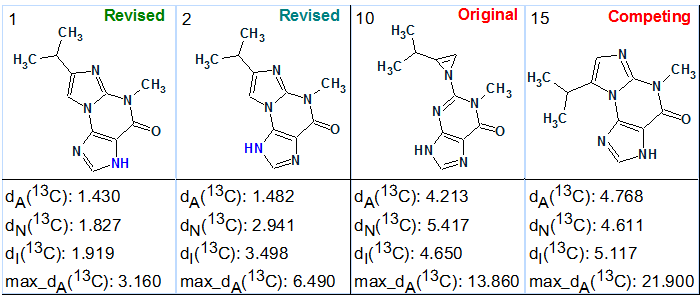
Figure 4. Selected structures of the ranked output file.
The revised structure 2 (5a) is ranked second, while its tautomer had minimal calculated deviations and was placed in the first position. The original structure 1 was placed in 10th position with a difference in calculated chemical shift deviations the value of deviation difference D = d(10) – d(1) ~ 3 ppm. Therefore the wrong original structure 1 was reliably rejected by Structure Elucidator Suite. The competing regioisomer 5b of the correct structure was placed in 15th position and also reliably rejected by the program.
Thus, our investigation showed convincingly that application of ACD/Structure Elucidator Suite could determine the correct acremolin structure with high reliability, almost instantly and without application of 1H–15N HMBC data. If the program was used from the very beginning, structures 1 and 5b hardly would attract attention of the original researchers [1] due to the very large average deviations calculated for them. Moreover, large and laborious work—the five-step synthesis of the revised structure—would be unnecessary for the authors [2], saving the time of highly qualified chemists.
For completeness, we added the 1H–15N HMBC correlations shown in Figure 2 to the set of experimental data and created a new MCD (Figure 5).
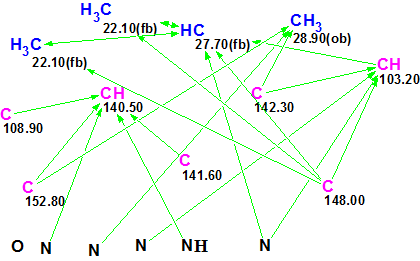
Figure 5. New Molecular Connectivity Diagram. Connectivities corresponding to 1H–13C and 1H–15N HMBC are shown.
Fuzzy Structure Generation from this new MCD gave the following results: k=67→65→55, tg = 0.5 s. As expected, the additional structural information carried by H–N HMBC led to a dramatic reduction in both the number of generated structures and processing time (from 100 sec to 0.5 sec). The first two structures shown in Figure 4 and competing regioisomer 3 were again ranked in the same order, but the original structure 1 was not generated because H–N HMBC correlations contradict it. The structures of regioisomer 3 with 15N experimental and calculated (3a) chemical shifts and connectivities are displayed below:
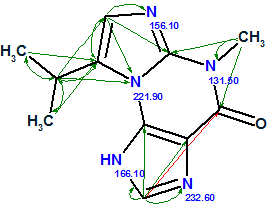 3 |
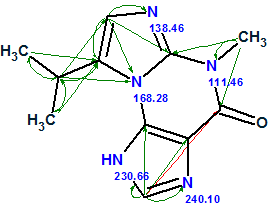 3a |
We see that structure 3 can be rejected not only by its large average 13C deviations (~5 ppm), but also by the large deviations calculated for nitrogen atoms N 221.9 and N 166.1.
We have demonstrated how the acremolin structure was determined using standard CASE methodology. However we can also examine the simplest way to disprove the original structure 1 and find the correct one, using the command Structure/Create Project for Structure. The command was applied after drawing structure 1, where 13C and 1H chemical shifts were assigned. As a result an artificial Molecular Connectivity Diagram was created (Figure 6):
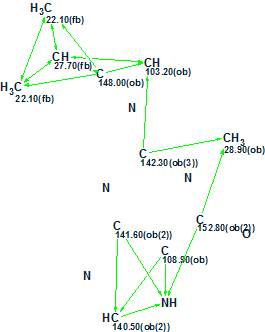
Figure 6. Artificial MCD created from structure 1.
In this MCD, all theoretically possible HMBC correlations were drawn by the program. The question was posed: which structures except structure 1 will be generated from the artificial MCD? Structure generation gave the following results: k = 96 → 4, tg = 0.06 s, and the ranked output file is presented in Figure 7.
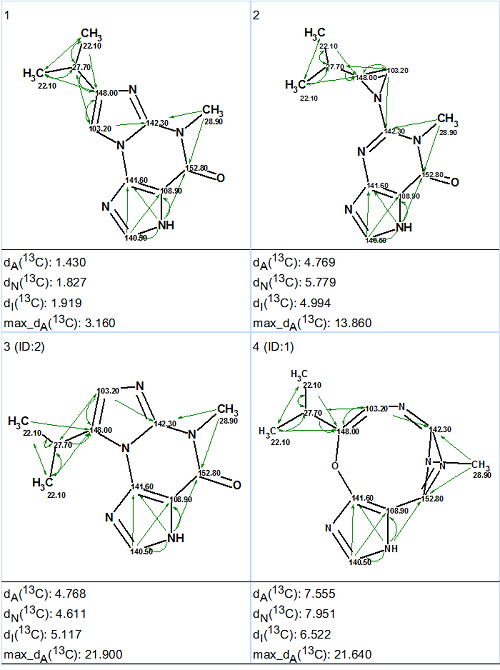
Figure 7. The ranked output file generated from the artificial MCD. All theoretically possible connectivities are shown.
Figure 7 gives answers to all of the questions which were posed by Januar and Molinski [2]. The complete solution to the problem was found in a second without any synthesis and additional NMR experiments, as well as without the application of standard CASE methodology.
We conclude that although the classic dictum “synthesis is the ultimate proof of structure” continues to work, it should be complemented by the following statement: before starting total synthesis for structure revision it is very desirable to take into account results delivered by a CASE system.
References
- Julianti, E.; Oh, H.; Lee, H.-S.; Oh, D.-C.; Oh, K.-B.; Shin, J. (2012) Acremolin, a new 1H-azirine metabolite from the marine-derived fungusAcremonium strictumTetrahedron Lett., 53:2885–2886.
- Januar, L. A.; Molinski T. F. (2013) Acremolin from Acremonium strictum is N2,3-Etheno-20-isopropyl-1-methylguanine, not a 1HAzirine. Synthesis and Structural Revision. Org. Lett., 15(10):2370–2373.
- Banert, K. (2012) Acremolin, a stable natural product with an antiaromatic 1H-azirine moiety? A structural reorientation. Tetrahedron Lett., 53:6443–6445.


