November 1, 2012
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Aetheramide
Aetheramides are structurally distinctive cyclic peptides isolated from a novel myxobacterial genus with the proposed name of “Aetherobacter”. These natural products are characterized by both their unique structures and their potential for HIV inhibition. Plaza et al [1] isolated two new unusual depsipeptides, aetheramides A and B. Their structures were elucidated using extensive NMR (1H, 13C, HSQC, COSY, HMBC, HOHAHA) and ESIMS analysis, chemical derivatizations, and by the combined analysis of homonuclear (H-H) and heteronuclear 2,3JCH-couplings and quantum mechanical calculations. Here we will discuss the computer-assisted structure elucidation (CASE) of aetheramide B (1) using ACD/Structure Elucidator.
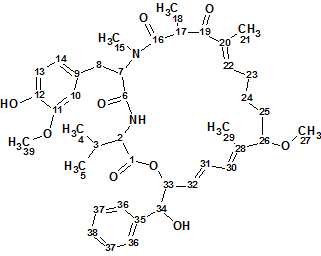
Structure 1
The HRESIMS of aetheramide B displayed an [M+ H]+ peak at m/z 719.3920, consistent with the molecular formula C41H54N2O9 (with a calculated mass of 719.3908). The degree of unsaturation is equal to 16. The IR spectrum displayed absorption bands at 3367 and 1673 cm-1, suggesting the presence of OH and/or NH (3367 cm-1), and carbonyl and/or olefin functionalities. The NMR spectroscopic data used for CASE analysis are presented in Table 1.
Table 1. NMR spectroscopic data used for CASE.
| C 1 | 169.4 | 171.09 | C | ||||
| C 2 | 58.8 | 57.91 | CH | 3.92 | u | 170.30, 18.20, 169.40, 18.60, 29.00 | |
| C 3 | 29 | 31.05 | CH | 1.91 | u | 18.60, 58.80, 18.20, 169.40 | |
| C 4 | 18.6 | 17.58 | CH3 | 0.56 | d(6.8) | 18.20, 58.80, 29.00 | |
| C 5 | 18.2 | 18.65 | CH3 | 0.78 | d(6.8) | 18.60, 29.00, 58.80 | |
| C 6 | 170.3 | 169.9 | C | ||||
| C 7 | 54.9 | 56.29 | CH | 5.5 | s | 35.20, 171.70, 31.10, 127.30, 170.30 | |
| C 8 | 35.2 | 35.62 | CH2 | 3.07 | u | 112.50, 170.30, 54.90, 127.30, 120.90 | |
| C 8 | 35.2 | 35.62 | CH2 | 2.77 | u | ||
| C 9 | 127.3 | 130.26 | C | ||||
| C 10 | 112.5 | 111.82 | CH | 6.77 | u | 120.90, 114.50, 35.20, 127.30, 144.60, 146.90 | |
| C 11 | 146.9 | 146.34 | C | ||||
| C 12 | 144.6 | 145.24 | C | ||||
| C 13 | 114.5 | 114.85 | CH | 6.6 | d(8) | 127.30, 146.90 | |
| C 14 | 120.9 | 122.05 | CH | 6.59 | d(8) | 144.60, 35.20, 112.50 | |
| C 15 | 31.1 | 31.49 | CH3 | 2.96 | s | 171.70, 54.90 | |
| C 16 | 171.7 | 171.19 | C | ||||
| C 17 | 42.7 | 45.86 | CH | 4.24 | u | 12.10, 197.20, 171.70 | |
| C 18 | 12.1 | 13.31 | CH3 | 0.58 | d(6.7) | 197.20, 42.70, 171.70 | |
| C 19 | 197.2 | 197.54 | C | ||||
| C 20 | 135.2 | 137.3 | C | ||||
| C 21 | 11.3 | 10.09 | CH3 | 1.65 | s | 140.40, 135.20, 197.20 | |
| C 22 | 140.4 | 144.7 | CH | 6.45 | u | 2.17 | 23.30, 11.30, 197.20, 27.10 |
| C 23 | 27.1 | 29.03 | CH2 | 2.17 | u | 6.45, 1.11 | 135.20, 31.40, 140.40, 23.30 |
| C 23 | 27.1 | 29.03 | CH2 | 1.99 | u | ||
| C 24 | 23.3 | 24.25 | CH2 | 1.11 | u | 1.42, 2.17 | 140.40, 27.10, 31.40, 84.90 |
| C 25 | 31.4 | 32.11 | CH2 | 1.42 | u | 3.48, 1.11 | 84.90, 138.70, 27.10, 23.30 |
| C 25 | 31.4 | 32.11 | CH2 | 1.55 | u | 138.70, 126.70, 84.90 | |
| C 26 | 84.9 | 80.54 | CH | 3.48 | u | 1.42 | 31.40, 138.70, 23.30, 126.70, 55.10 |
| C 27 | 55.1 | 55.52 | CH3 | 3.03 | u | 84.9 | |
| C 28 | 138.7 | 134.66 | C | ||||
| C 29 | 9.6 | 19.45 | CH3 | 1.55 | s | 138.70, 126.70, 84.90 | |
| C 30 | 126.7 | 129.1 | CH | 5.8 | d(11.2) | 6.44 | 84.90, 9.60, 138.70, 127.50, 130.20 |
| C 31 | 130.2 | 133.42 | CH | 6.44 | u | 5.80, 5.60 | 78.50, 138.70, 126.70, 127.50 |
| C 32 | 127.5 | 130.12 | CH | 5.6 | u | 5.38, 6.44 | 138.70, 73.10, 78.50, 126.70 |
| C 33 | 78.5 | 79.31 | CH | 5.38 | u | 5.60, 4.77 | 169.40, 73.10, 130.20, 141.30, 127.50 |
| C 34 | 73.1 | 74.65 | CH | 4.77 | u | 5.38 | 127.50, 141.30, 126.60, 78.50 |
| C 35 | 141.3 | 138.66 | C | ||||
| C 36 | 126.6 | 126.32 | CH | 7.39 | d(7.5) | 73.10, 127.60, 127.10 | |
| C 37 | 127.6 | 127.93 | CH | 7.32 | t(7.5) | 127.10, 126.60, 141.30 | |
| C 38 | 127.1 | 128.88 | CH | 7.23 | t(7.5) | 126.60, 141.30 | |
| C 39 | 55.2 | 55.99 | CH3 | 3.71 | u | 146.9 | |
| N 1 | 100* | NH | 8.13 | u | 29.00, 58.80, 169.40 | ||
| O 1 | 50* | OH | 8.67 | u |
*Chemical shifts of nitrogen and oxygen atoms (marked by *) are conventionally assigned to formally distinguish between different heteroatoms of the same type.
Molecular Connectivity Diagram (MCD) overview. The software automatically generated a molecular connectivity diagram from the experimental data (Figure 1). The degree of unsaturation is fairly large (16) but the molecular structure produces a large number of detected HMBC correlations. Four carbonyl bonds are evident and two O-CH3 bonds were drawn by hand to reduce the time required for structure generation. Carbon atoms with 13C chemical shifts in the range 54–85 ppm were supplied with the property sp3/ob (obligatory, i.e., must be sp3), which was supported by the corresponding 1H chemical shifts (see Table 1). Since the neighborhood of a nitrogen atom can vary the chemical shifts of sp2 carbons over a wide interval, the properties of the carbon atoms at 112.5–146.9 were marked as sp2/nd (not defined, i.e. the neighboring atom is undefined). The methyl group CH3 (δC 31.10; δH 2.96) is most likely chemically bonded to a nitrogen atom, but there are two nitrogen atoms in the molecule and it is not known which of them should be chosen as the associated atom, hence the methyl group was left without any chemical bond.
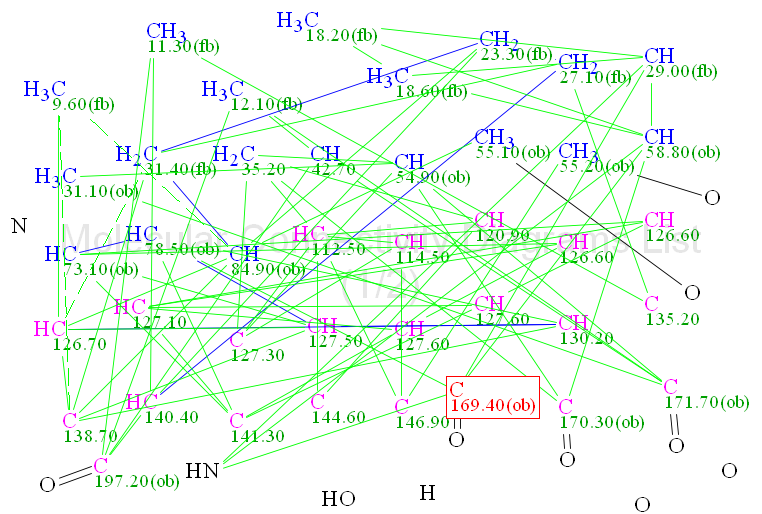
Figure 1. MCD with two O-CH3 bonds added by hand.
Checking the MCD consistency with the software revealed the presence of at least one non-standard connectivity. Taking this fact into account, fuzzy structure generation (FSG) was initiated using the mode “Determine option automatically” accompanied with 13C chemical shift prediction and structure filtering. The result gave: k = 53 → 6 → 2, tg = 22m 10s, 3 of the 75 correlations were extended during the generation process and 12785 from a total 67525 (~20%) possible fuzzy combinations were used. The structures elucidated are shown in Figure 2.
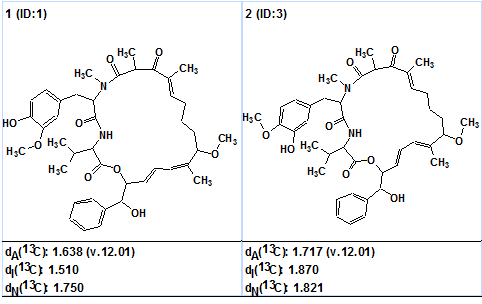
Figure 2. Ranked output structural file.
A comparison with Structure 1 shows that the structure #1 ranked as the first by all three methods is aetheramide B. The differences between deviations related to structures #1 and #2 are small because both structures are very similar: they differ only in the positions of the OH and CH3 groups on the benzene ring. Difficulties associated with the presence of three nonstandard correlations in the HMBC data were overcome by the program in automatic mode. The structure of aetheramide B (1a) with the chemical shift assignments and non-standard correlations included (colored in green) is shown below:
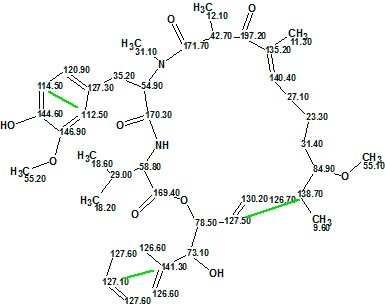
Structure 1a
Comparison of the experimental and calculated 13C chemical shifts (listed in Table 1) leads to the conclusion that chemical shift prediction performed very well.
This problem is another example that demonstrates the effectiveness of employing multiplicities in 1H NMR when they appear reliable (see also the previous Elucidation of the Month). When the multiplicities shown in the Table 1 were used for setting numbers of hydrogen atoms attached to neighbor carbons, the following result was obtained: k = 35 → 2 → 1, tg = 14m 45s. Only a single structure was output by the program, which was the correct structure (matching Structure 1), while the time of structure generation was decreased by ~ 30%.
References
- Aetheramides A and B, Potent HIV-Inhibitory Depsipeptides from a Myxobacterium of the New Genus “Aetherobacter”. A. Plaza, R. Garcia, G. Bifulco, J. P. Martinez, S. Hüttel, F. Sasse, A. Meyerhans, M. Stadler, R. Müller. Org. Lett., 14 (11):2854–2857, 2012. DOI: 10.1021/ol3011002


