December 1, 2013
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Ascidia sydneiensis SAAF
A Novel Sperm-Activating and Attracting Factor (SAAF)
Sperm activation and chemotaxis (a gradient of chemoattractant that guides sperm to an egg) ubiquitously occurs in animal species, including humans. In some situations, a single agent can induce both the sperm activation and attraction functions. To understand the molecular mechanism underlying the genus-specificity of ascidians’ sperm chemotaxis, Matsumori et al [1] investigated the structure of a novel compound Ascidia-SAAF 2 (1), which was isolated from the eggs of the ascidian Ascidia sydneiensis.
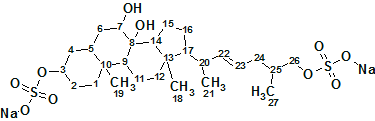
1
4 nmol (2.6 μg) of Ascidia-SAAF 2 was isolated from the active HPLC fractions. The chemical structure of Ascidia-SAAF 2 was elucidated using NMR and MS spectroscopy data. The molecular formula of Ascidia-SAAF 2, C27H44O10S2Na2, was obtained from the negative-ion high-resolution MS (m/z 296.1188, [M-2Na]2-, calculated m/z 296.1193), indicating a dehydrogenated or oxygenated (-H2) form of the known compound Ciona-SAAF 1 (C27H46O10S2Na2).
The gDQF-COSY and gHMBC spectra obtained using the cold-probe technology unambiguously demonstrated the presence of a double bond between C-22 and C-23. Although chemical shifts of H-22 and H-23 almost overlapped to provide second-order signals, its E configuration was identified on the basis of a large 3JHH (17 Hz) between H-22 and H-23, deduced from a spectral simulation of the signals. The absence of ROE between H-21 and H-24 could lend support for the presence of this configuration.
The spectroscopic NMR data (1H, 13C, HSQC, COSY and HMBC) used for the molecular structure elucidation are presented in Table 1.
Table 1. Ascidia-SAAF 2 spectroscopic NMR data. (click to expand)
| Label | δC | δC calc |
CHn | δH | M(J) | COSY | C HMBC |
| C 1 | 33.3 | 37.13 | CH2 | 1.18 | u | 1.90, 3.56 | |
| C 1 | 33.3 | 37.13 | CH2 | 1.55 | u | 1.84 | |
| C 2 | 26.5 | 26.54 | CH2 | 1.8 | u | ||
| C 2 | 26.5 | 26.54 | CH2 | 1.84 | u | 1.55, 4.65 | |
| C 3 | 78.5 | 81.9 | CH | 4.65 | u | 1.60, 1.84 | |
| C 4 | 18.0 | 34.16 | CH2 | 1.6 | u | 1.90, 4.65 | |
| C 4 | 18.0 | 34.16 | CH2 | 1.55 | u | ||
| C 5 | 32.6 | 39.37 | CH | 1.9 | u | 1.18, 1.60 | |
| C 6 | 31.5 | 32.04 | CH2 | 1.9 | u | ||
| C 6 | 31.5 | 32.04 | CH2 | 1.18 | u | ||
| C 7 | 72.0 | 72.4 | CH | 3.56 | u | 1.18 | |
| C 8 | n.d. | 76.14 | C | ||||
| C 9 | 50.2 | 51.14 | CH | 1.25 | u | 1.59 | |
| C 10 | 36 | 37.68 | C | ||||
| C 11 | 32.9 | 20.04 | CH2 | 1.59 | u | 1.21, 1.25 |
|
| C 12 | 40.7 | 37.06 | CH2 | 1.21 | u | 1.59 | |
| C 12 | 40.7 | 37.06 | CH2 | 1.99 | u | ||
| C 13 | 43.2 | 42.42 | C | ||||
| C 14 | 53.9 | 54.35 | CH | 1.54 | u | 1.38 | |
| C 15 | 18.6 | 24.3 | CH2 | 1.47 | u | ||
| C 15 | 18.6 | 24.3 | CH2 | 1.38 | u | 1.28, 1.54 | |
| C 16 | 28.4 | 28.43 | CH2 | 1.28 | u | 1.14, 1.38 | |
| C 16 | 28.4 | 28.43 | CH2 | 1.73 | u | ||
| C 17 | 56.6 | 55.04 | CH | 1.14 | u | 2.07, 1.28 | |
| C 18 | 13.5 | 20.57 | CH3 | 0.9 | u | C 13, C 12, C 9, C 17, C 14, C 1, C 10, C 5 |
|
| C 19 | 11.5 | 21.58 | CH3 | 0.9 | u | ||
| C 20 | 39.7 | 32.67 | CH | 2.07 | u | 0.98, 1.14, 5.39 | |
| C 21 | 20.3 | 20.85 | CH3 | 0.98 | u | 2.07 | C 22, C 20, C 17 |
| C 22 | 140.5 | 140.59 | CH | 5.39 | u | 2.02, 2.07 | |
| C 23 | 125.5 | 128.38 | CH | 5.39 | u | ||
| C 24 | 36.0 | 33.32 | CH2 | 1.93 | u | C 22, C 23 | |
| C 24 | 36.0 | 33.32 | CH2 | 2.02 | u | 5.39, 1.87 | |
| C 25 | 33.5 | 34.07 | CH | 1.87 | u | 0.92, 2.02, 3.83 | |
| C 26 | 74.1 | 76.27 | CH2 | 3.83 | u | 1.87 | |
| C 26 | 74.1 | 76.27 | CH2 | 3.95 | u | ||
| C 27 | 16.4 | 17.41 | CH3 | 0.92 | u | 1.87 | C 26, C 24, C 25 |
Using this data, a Molecular Connectivity Diagram (MCD) was created in ACD/Structure Elucidator Suite, and is presented in Figure 1.
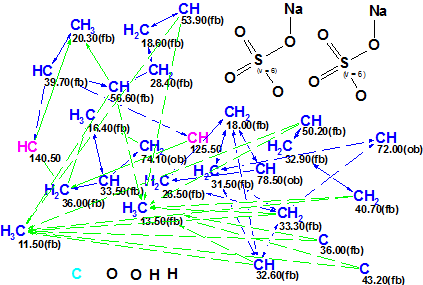
Figure 1. Ascidia-SAAF 2 Molecular Connectivity Diagram.
MCD overview. Two SO4Na fragments were manually added as their presence was postulated by authors [1] from previous chemical knowledge. The 13C NMR spectrum appeared to be missing one carbon atom signal, so it was introduced in Table 1 without any chemical shift (C 8). This atom is likely a quaternary one, and it was marked manually as “sp2 or sp3“-hybridized on the MCD (light blue colored). One can see from Table 1 that there are three pairs of overlapped 1H signals (italic underlined), and this is a reason for the large number of ambiguous connectivities (marked by dotted lines in the MCD) making the 2D NMR data less definite. At the same time, nine carbon atoms are supplied with labels “fb” defining their admissible neighbors (neighboring with heteroatoms is forbidden), which accelerates the structure generation significantly and reduces size of the output structural file.
No contradictions were detected in 2D NMR data by the software, and Strict Structure Generation was initiated, combined with 13C chemical shift prediction and utilizing an average deviation threshold (d=4 ppm). Results: k=2880→20→8, tg=31.5s. The two top structures of the ranked output file are presented in Figure 2.
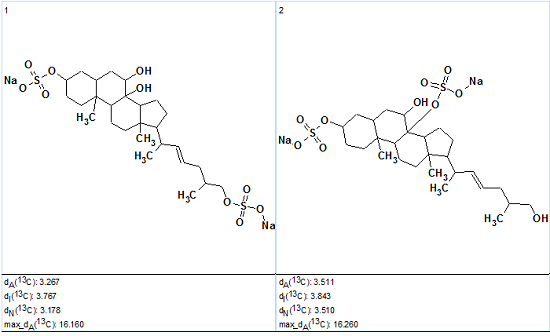
Figure 2. Two top structures of the ranked output file for Ascidia-SAAF 2.
Figure 2 shows that the top-ranked structure coincides with the structure of Ascidia-SAAF 2, though the difference D=d(2)-d(1) is small and values of deviations are relatively large, making conclusive verification less definite. However these values can be explained through comparison of the experimental 13C chemical shifts with predicted ones. Table 1 shows that the largest discrepancies between experimental and calculated values are observed for atoms C4 and C11 (selected by red color). The Chemical Shift Calculation protocols generated for these atoms are presented in Figures 3 and 4 correspondingly.
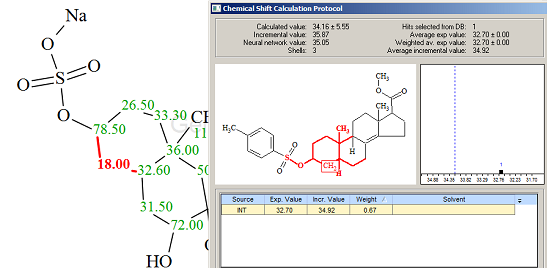
Figure 3. Chemical Shift Calculation Protocol for the carbon atom C 4 (δ18.0) of Ascidia-SAAF 2.
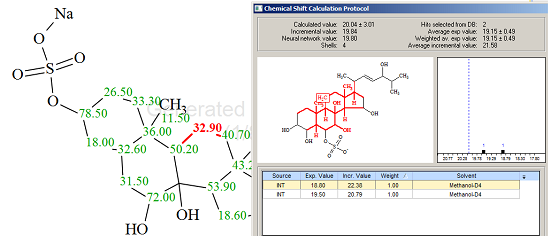
Figure 4. Chemical Shift Calculation Protocol for the carbon atom C 11 (δ32.9) of Ascidia-SAAF 2.
Only one hit was found in the first case, and two hits in the second, but these results allows one to suggest that chemical shifts 18.00 and 32.90 ppm should be exchanged. When the exchange was carried out the average deviations reduced to the following values: dA=2.278, dI=2.864 and dN=2.170 ppm. We believe that this result can be considered as justification of the atom permutation.
As was established by authors [1] on the basis of a large 3JHH (17 Hz) between H-22 and H-23, the single double bond C=C exists in the original structure 1 in E configuration. However 13C chemical shift prediction using ACD/NMR Predictors can also help in distinguishing between E and Z configurations. When the ranked structure #1 was transferred into Z configuration by moving substituents upon the double bond, the deviations increased to dA=2.584, dI=3.079 and dN=2.422 ppm.
Therefore utilizing Structure Elucidator Suite allowed the correct structure of unknown, and its configuration upon a double bond to be determined.
The elucidated structure 1a with refined chemical shift assignment is shown below:
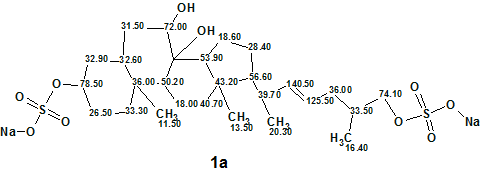
1a
As the experimental 13C chemical shift value was not determined for the carbon C 8, its approximate value can be assigned as 76 ppm on the basis of 13C chemical shift prediction (see Table 1).
References
- N. Matsumori, Y. Hiradate, H. Shibata, T.Oishi, S. Shimma, M.Toyoda, F. Hayashi, M. Yoshida, M.Murata, M. Morisawa, A Novel Sperm-Activating and Attracting Factor from the Ascidian Ascidia Sydneiensis. Org. Lett., 15(2):294–297, 2013.


