September 1, 2021
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Davidone F
Sophora davidii (Franch.) belong to the family Fabaceae and are shrubs or small trees found in China. Over the years many natural products have been isolated from them, including alkaloids, steroids, lignans, phenolic acids and, mostly, flavonoids. These have been found to have varying biological activities.
The chemical investigation of S. davidii which was performed by Song et al [1] led to the isolation of a new prenylated flavanone, davidone F (1), with a novel seven-membered oxygen ring.
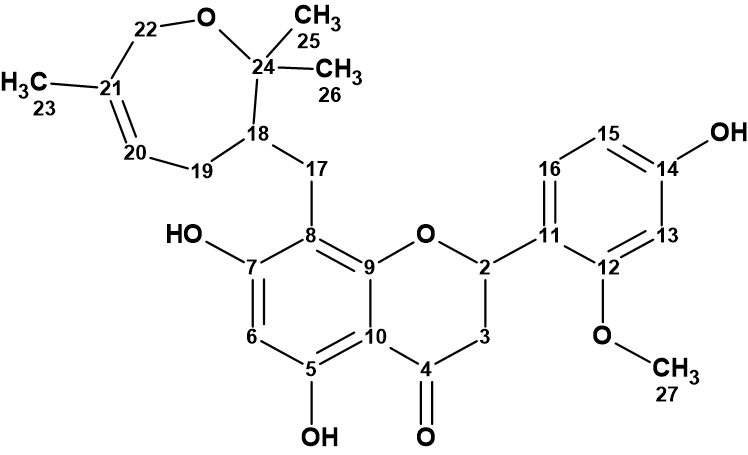
1
Compound 1 was isolated as a brown oil. The molecular formula was found to be C26H30O7 by HR-ESI-MS data (m/z 455.2061 [M + H]+, calculated for 455.2064), indicating 12 indices of hydrogen deficiency. The IR spectrum showed the characteristic absorption bands of hydroxy (3244 cm-1) and conjugated carbonyl (1636 cm-1) groups.
The structure of davidone F was determined using 1D and 2D (HSQC, HMBC and COSY) NMR spectra. These data were used to challenge ACD/Structure Elucidator. It should be noted that only the key HMBC and COSY correlations were listed in the publication [1], not the complete list (see Table 1).
Table 1. Spectroscopic NMR data of davidone F.
| Label | δC | δC calc (HOSE) | CHn | dH | COSY | H to C HMBC |
| C 2 | 75.500 | 73.970 | CH | 5.610 | 3.13 | C 11, C 12, C 9, C 4 |
| C 3 | 42.700 | 42.500 | CH2 | 3.130 | 5.61 | C 2, C 4 |
| C 3 | 42.700 | 42.500 | CH2 | 2.660 | ||
| C 4 | 198.700 | 197.330 | C | |||
| C 5 | 163.300 | 160.700 | C | |||
| C 6 | 96.300 | 97.560 | CH | 5.940 | C 10, C 8, C 5, C 7 | |
| C 7 | 166.300 | 162.110 | C | |||
| C 8 | 108.200 | 108.310 | C | |||
| C 9 | 162.500 | 161.090 | C | |||
| C 10 | 103.300 | 102.170 | C | |||
| C 11 | 119.200 | 118.900 | C | |||
| C 12 | 159.300 | 161.240 | C | |||
| C 13 | 99.800 | 98.900 | CH | 6.480 | C 15, C 11, C 12, C 14 | |
| C 14 | 160.500 | 157.300 | C | |||
| C 15 | 108.000 | 107.200 | CH | 6.430 | 7.32 | C 14 |
| C 16 | 129.100 | 127.400 | CH | 7.320 | 6.43 | C 2, C 12, C 14 |
| C 17 | 26.000 | 26.240 | CH2 | 2.200 | 2.40,1.75 | C 24 |
| C 17 | 26.000 | 26.240 | CH2 | 2.400 | 2.20 | C 19, C 8, C 9, C 7 |
| C 18 | 48.900 | 48.140 | CH | 2.200 | ||
| C 19 | 28.500 | 28.620 | CH2 | 2.180 | ||
| C 19 | 28.500 | 28.620 | CH2 | 1.750 | 2.20,5.29 | C 24, C 20, C 21 |
| C 20 | 126.600 | 122.970 | CH | 5.290 | 1.75 | |
| C 21 | 136.800 | 136.060 | C | |||
| C 22 | 65.700 | 65.180 | CH2 | 4.260 | C 24, C 20 | |
| C 22 | 65.700 | 65.180 | CH2 | 3.680 | ||
| C 23 | 20.900 | 20.920 | CH3 | 1.520 | C 22, C 20, C 21 | |
| C 24 | 80.300 | 79.530 | C | |||
| C 25 | 24.300 | 24.370 | CH3 | 1.180 | C 18, C 24 | |
| C 26 | 22.500 | 24.210 | CH3 | 1.080 | C 18, C 24 | |
| C 27 | 55.900 | 55.200 | CH3 | 3.810 | C 12 |
The data presented in Table 1 were entered into ACD/SE and a Molecular Connectivity Diagram (MCD) was automatically created by the program (Figure 1).
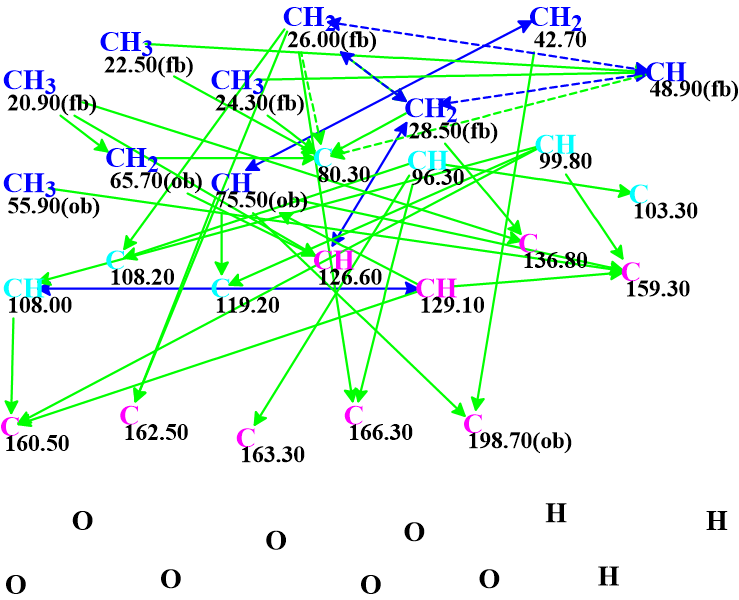
Figure 1. Molecular connectivity diagram.
MCD overview. The MCD contains seven carbon atoms colored light blue, with chemical shifts in the range from 80.3 up to 119.2 ppm. For these atoms the ambiguous hybridization not sp (i.e. sp2 or sp3) was set by the program. This means that both possible hybridizations should be explored for each mentioned carbon atom during structure generation. This usually increases the elucidation time. The dotted blue lines indicate ambiguous COSY correlations which appeared due to the presence of two overlapping signals at 2.20 ppm in the 1H NMR spectrum (marked red in Table 1).
No user edits were made to the MCD. Structure generation together with incremental 13C chemical shift prediction was initiated, and the following results were obtained: k = 1288 → (structural and spectral filtering) → 5, tg = 16s.
According to CASE methodology, 13C chemical shift calculation was performed for the 5 structures in the output file using the three methods implemented into ACD/SE (incremental, neural networks and HOSE code based). The structures were ranked in order of increasing average deviations of the calculated 13C chemical shifts from experimental ones. The final output file, with the structures ranked, is shown in Figure 2.
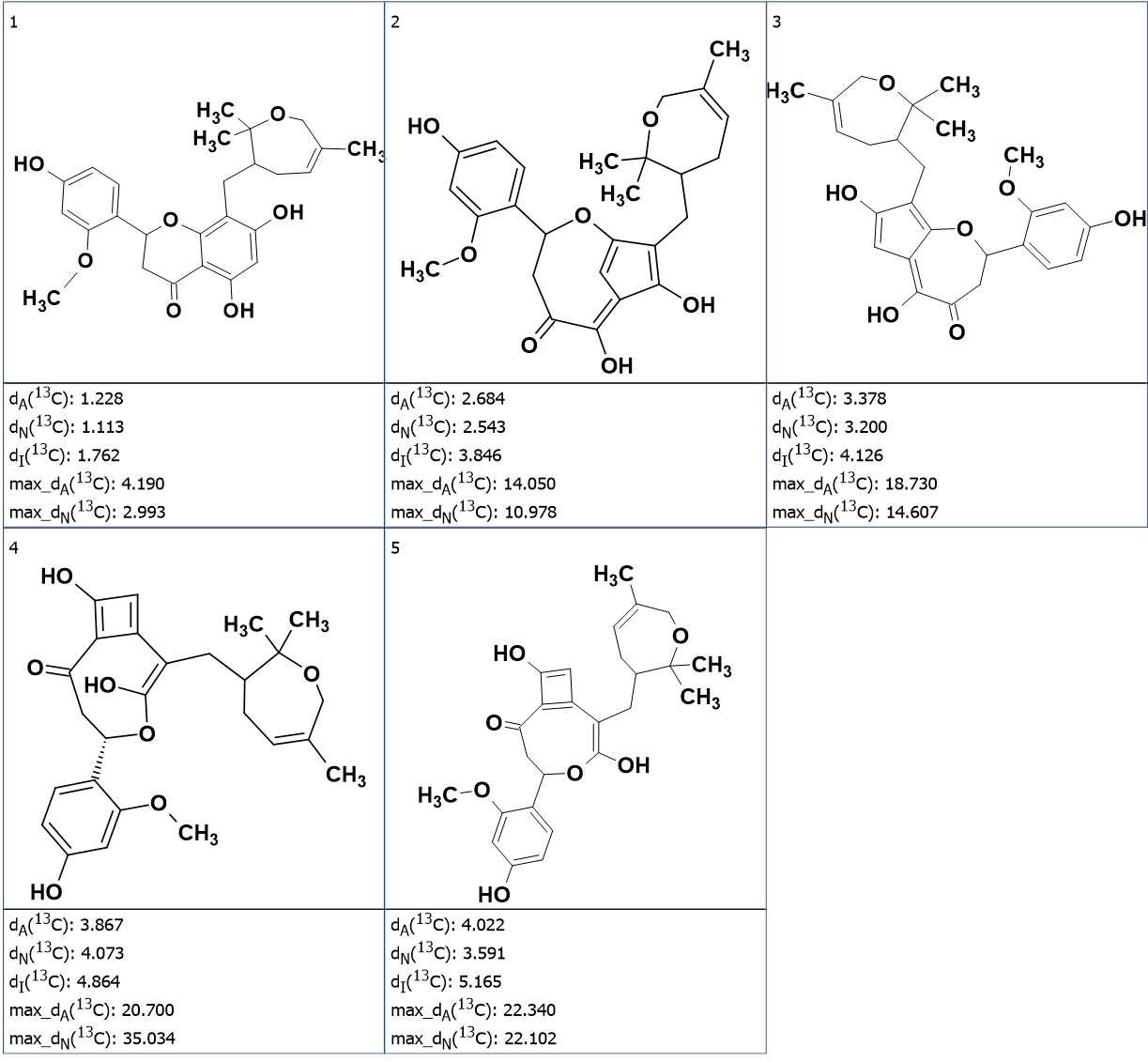
Figure 2. The final output file.
We see that the first structure coincides with the one determined in the publication [1]. Small values of average deviations allow us to conclude that the elucidation of the structure was reliable, and no additional calculations (e.g. DFT chemical shift predictions, DP4 probabilities) are required. It is interesting to note that the solution to the problem was found very quickly, in 16 s, despite the presence of seven carbon atoms with ambiguous hybridizations. This was possible due to the implementation of multi-threaded structure generation algorithms in the program, essentially splitting the initial MCD in several others, each exploring a single case of the possible hybridizations combinations, and all solved in parallel using a multi-core CPU.
According to our experience ACD/SE can solve problems when only the HMBC data are used. We have seen in the past the structure generation time being reduced by a factor of 3 just by manually defining a carbonyl, and then by an additional factor of 30 by defining that a carbon atom must be connected to a heteroatom. This shows that adding even minimum evident structural constraints to the information displayed in the MCD can drastically accelerate the structure generation. Of course, in each case the effect will be different.
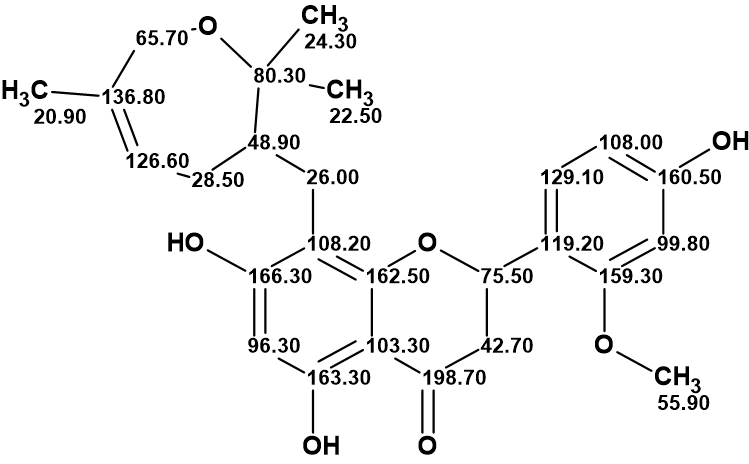
The structure of davidone F with the 13C chemical shift assignment as defined by the program is shown below.
References
- Davidones F and G, Two Novel Flavonoids from Sophora davidii (Franch.) Skeels P.Song, X. Li , T. Zhou, Y. Peng, H.-Y. Choi, Y. Ma, X. Yang. (2021). Molecules, 26, 4182. https://doi.org/10.3390/


