April 1, 2017
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Delicoferone A
The fungi that colonize and reproduce in animal excrements are called Coprophilous fungi. They have specifically adapted to colonize in that environment, however, they can also be found on plants and soils when their spores are dispersed there.
Jayanetti at co-workers [1] investigated cultures of such a fungus, the ex-type strain of Delitschia confertaspora (ATCC 74209). This strain was part of the 1000 Fungal Genomes Project, and its genome has been sequenced as part of that project. It was shown to contain 30 or more gene clusters consistent with secondary metabolite biosynthetic processes. Chemical studies of fermentation extracts of the fungus produced three new benzophenone derivatives with a distinctive architecture. The structures of these metabolites were solved by using 2D NMR and HRES-TOFMS data, as well as by comparison with related compounds described in the literature. Spectroscopic data acquired for one of the new compounds, Delicoferone A (1), were used to challenge ACD/Structure Elucidator.
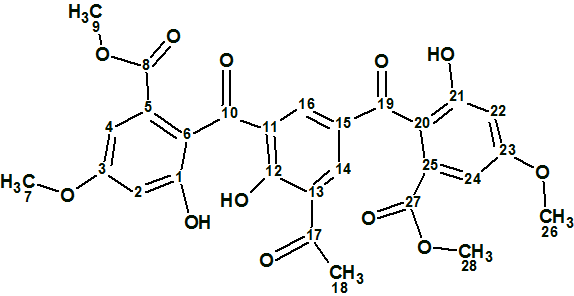
1
Delicoferone A (1) was isolated as a yellow solid. The molecular formula was determined to be C28H24O12 (17 unsaturations) on the basis of HRESITOFMS, m/z 553.1326 [M + H]+ (calcd. for C28H24O12+H, 553.1338), and NMR data. IR absorption bands observed at 3417 (br), 1722, 1670, 1650, 1608, and 1498 cm-1 indicate the presence of hydroxyl, ester and conjugated ketone groups as well as benzene rings. The NMR data extracted from 1H, 13C, HSQC and HMBC spectra, as well as the predicted 13C chemical shifts, are collected in Table 1.
Table 1. Delicoferone A. Spectroscopic NMR data
| Label | δC | δC calc* | CHn | δH | M/J | C to H HMBC |
| C 1 | 157.8 | 164.82 | C | |||
| C 2 | 106.5 | 103.72 | CH | 6.61 | d, 2.4 | C 1, C 3, C 4, C 6, C 10 |
| C 3 | 163.2 | 163.14 | C | |||
| C 4 | 107.9 | 104.73 | CH | 7.01 | d, 2.4 | C 2, C 3, C 5, C 6, C 8, C 10 |
| C 5 | 132.1 | 133.44 | C | |||
| C 6 | 121.2 | 125.64 | C | |||
| C 7 | 56.2 | 55.91 | CH3 | 3.83 | s | C 3 |
| C 8 | 167.5 | 167.13 | C | |||
| C 9 | 53 | 52.13 | CH3 | 3.65 | s | C 8 |
| C 10 | 202.3 | 199.61 | C | |||
| C 11 | 125 | 120.23 | C | |||
| C 12 | 166.9 | 167.2 | C | |||
| C 13 | 127 | 126.3 | C | |||
| C 14 | 139.6 | 131.37 | CH | 7.83 | d, 2.4 | C 12, C 13, C 15, C 16, C 17, C 19 |
| C 15 | 130.5 | 132.83 | C | |||
| C 16 | 137.6 | 136.85 | CH | 8.39 | d, 2.4 | C 10, C 11, C 12, C 14, C 15, C 19 |
| C 17 | 201.6 | 203.86 | C | |||
| C 18 | 30.9 | 29.44 | CH3 | 2.71 | s | C 13, C 17 |
| C 19 | 196.1 | 192.49 | C | |||
| C 20 | 121.8 | 126.03 | C | |||
| C 21 | 157.7 | 164.81 | C | |||
| C 22 | 106.6 | 102.92 | CH | 6.59 | d, 2.4 | C 19, C 20, C 21, C 23, C 24 |
| C 23 | 162.8 | 163.14 | C | |||
| C 24 | 107.8 | 105.1 | CH | 6.98 | d, 2.4 | C 19, C 20, C 22, C 23, C 25, C 27 |
| C 25 | 132.3 | 134.37 | C | |||
| C 26 | 56.2 | 55.91 | CH3 | 3.81 | s | C 23 |
| C 27 | 167.6 | 167.2 | C | |||
| C 28 | 52.9 | 52.13 | CH3 | 3.59 | s | C 27 |
*13C chemical shift prediction was performed by HOSE code based approach using ACD/CNMR Predictor.
The Molecular Connectivity Diagram (MCD) created from the data presented in Table 1, and edited as described below, is shown in Figure 1.
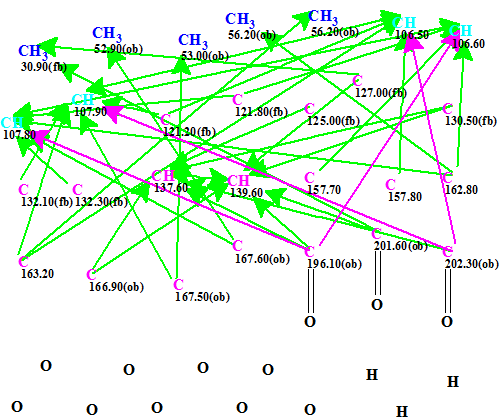
Figure 1. Molecular connectivity diagram.
MCD overview. As evident from the molecular formula, the compound contains 40 skeletal atoms from which 12 atoms are oxygens, while the degree of unsaturation is 17. Under these conditions it is not unrealistic to expect the user to help the program by introducing some additional information capable of reducing the time of structure generation and the size of the output file. Comparison of the list of HMBC correlations with the HMBC pattern presented in Supporting Information (Figure 3) allows one to conclude that intensities of the peaks corresponding to correlations H-2/C-10, H-4/C-10, H-22/C-19 and H-24/C-19 listed in [1] are very low (the peaks are almost invisible). Therefore the lengths of these correlations were set as 2-4 bonds (pink arrows in MCD). Hybridization of carbons 106.5, 106.6, 107.8 and 107.9 (marked by light blue color) was set by the program as not sp (sp3 or sp2), having in mind that O-CH-O fragment may present in the structure. Carbons C(121.2) – C(132.3) were assumed to be in sp2 hybridization state and neighboring with an oxygen atom is forbidden for them. Three evident carbonyl bonds were manually defined and multiplicities and coupling constants of all 1H signals (Table 1) were used to specify the number of hydrogens situated in the first sphere around corresponding carbons which bear hydrogens (all are zeroes in the given case) .
No contradictions were detected in the HMBC data and strict structure generation accompanied with 13C chemical shift prediction and structural filtering was initiated. Results: k=4076→131→47, tg =3 min.
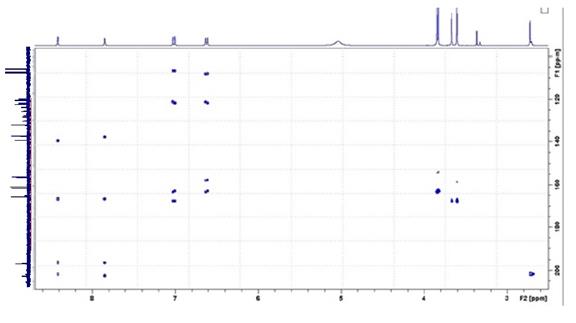
Figure 2. HMBC pattern.
13C chemical shift prediction was performed for all structures of the output file and the structures were ranked in ascending order of average deviations dA (13C chemical shifts were calculate by HOSE code based approach). The eight top structures of the ranked file are presented in Figure 3.
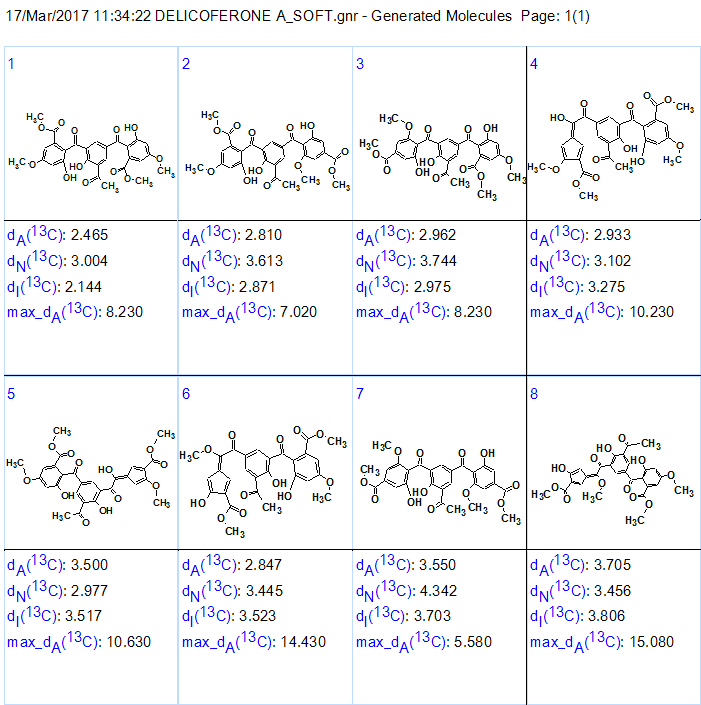
Figure 3. The top eight structures of the ranked output file.
We see that structure #1 is characterized with smallest average deviations calculated by all three methods of NMR spectrum prediction implemented into ACD/Structure Elucidator [2]. This structure is identical to the structure suggested for Delicoferone A.
The elucidated structure with automatically assigned 13C chemical shifts is shown below.
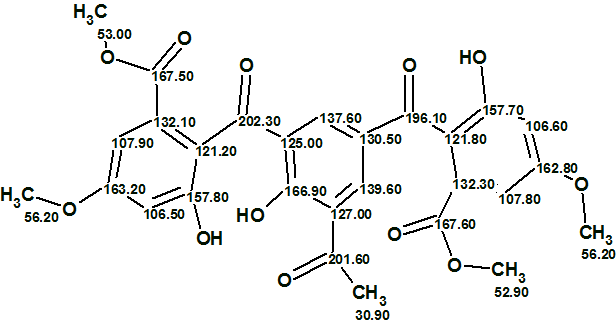
In conclusion, the structure of a highly complex oxygenated molecule was elucidated by NMR data alone in just 3 minutes with only very minor manual help.
References
- D. R. Jayanetti, Y. Li, G. A. Bartholomeusz, G. F. Bills, J.B. Gloer. (2017). Benzophenone and Fimetarone Derivatives from the Coprophilous Fungus Delitschia confertaspora. J. Nat. Prod., 80(3): 709–712. DOI: 10.1021/acs.jnatprod.6b01091.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.


