April 1, 2019
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
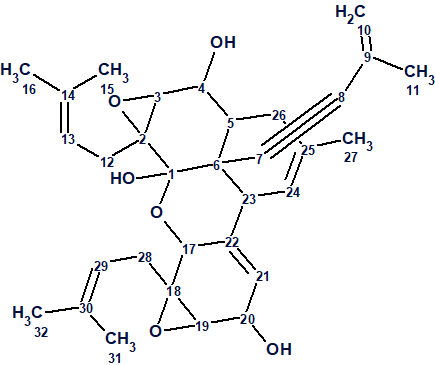
Dimericbiscognienyne A
Meroterpenoids are produced by the terpenoid biosynthetic pathway and have been studied extensively over the years. We have already presented some examples of Meroterpenoid structures being solved with Structure Elucidator (see, for example, Neosetophomone A, Manginoid A and 4-bromobenzoic biscognienyne A).
Zhao and coworkers [1] had been studying the secondary metabolites of Biscogniauxia sp, isolated from the Usnea mutabilis stirt. lichen. They isolated a diisoprenyl-cyclohexane-type meroterpenoid dimer with a new skeleton. Dimericbiscognienyne A 1 was isolated along with three related compounds.
1
Dimericbiscognienyne A was obtained as colorless block shaped crystals. The molecular formula of 1 was found to be C32H40O6 (13 degrees of unsaturation) from its HR-ESI-MS (m/z 543.2720 [M + Na]+, calcd for C32H40O6Na: 543.2723). The authors collected HSQC, HMBC and COSY spectra which are summarized in Table 1. Three pairs of carbons have identical 13C chemical shifts, and are marked in Table 1.
Table 1. Spectroscopic 1D and 2D NMR data of compound 1
| Label | δC | δC calc | XHn | δH | COSY | H to C HMBC |
| C 1 | 94.300 | 94.580 | C | |||
| C 2 | 64.400 | 63.320 | C | |||
| C 3 | 59.600 | 64.190 | CH | 3.050 | C 12, C 5, C 2, C 4 |
|
| C 4 | 66.900 | 71.190 | CH | 3.860 | 2.11 | C 26, C 5, C 3, C 2 |
| C 5 | 41.800 | 42.850 | CH | 2.110 | 3.86, 2.15 |
C 26, C 23, C 6, C 4, C 7, C 1, C 25 |
| C 6 | 48.300 | 48.930 | C | |||
| C 7 | 86.100 | 98.140 | C | |||
| C 8 | 88.200 | 99.270 | C | |||
| C 9 | 126.200 | 123.850 | C | |||
| C 10 | 122.100 | 123.020 | CH2 | 5.140 | ||
| C 10 | 122.100 | 123.020 | CH2 | 5.200 | C 11, C 8, C 9 |
|
| C 11 | 23.500 | 23.570 | CH3 | 1.780 | C 8, C 10, C 9 |
|
| C 12 | 26.200 | 29.210 | CH2 | 2.920 | ||
| C 12 | 26.200 | 29.210 | CH2 | 2.780 | C 3, C 2, C 13, C 14 |
|
| C 13 | 116.900 | 120.530 | CH | 5.020 | C 15, C 16 |
|
| C 14 | 135.600 | 136.680 | C | |||
| C 15 | 18.000 | 18.020 | CH3 | 1.630 | C 16, C 13, C 14 |
|
| C 16 | 25.800 | 25.770 | CH3 | 1.710 | C 15, C 31, C 13, C 29, C 14, C 30 |
|
| C 17 | 71.000 | 70.260 | CH | 4.740 | C 21, C 22 |
|
| C 18 | 60.200 | 61.910 | C | |||
| C 19 | 59.400 | 61.380 | CH | 3.210 | 4.45 | C 28, C 20, C 21 |
| C 20 | 64.100 | 65.190 | CH | 4.450 | 5.50,3.21 | C 18, C 21, C 22 |
| C 21 | 118.300 | 119.180 | CH | 5.500 | 4.45 | C 23, C 19, C 17 |
| C 22 | 134.700 | 130.340 | C | |||
| C 23 | 38.000 | 40.760 | CH | 3.910 | 5.40 | C 6, C 7, C 24, C 22 |
| C 24 | 117.700 | 124.810 | CH | 5.400 | 3.91 | C 27, C 26, C 23, C 6, C 22 |
| C 25 | 133.400 | 135.530 | C | |||
| C 26 | 31.200 | 30.920 | CH2 | 2.450 | ||
| C 26 | 31.200 | 30.920 | CH2 | 2.150 | 2.11 | C 27, C 5, C 6, C 4, C 24, C 25 |
| C 27 | 23.900 | 24.200 | CH3 | 1.730 | C 26, C 24, C 25 |
|
| C 28 | 31.300 | 31.720 | CH2 | 2.300 | ||
| C 28 | 31.300 | 31.720 | CH2 | 2.980 | C 19, C 18, C 17, C 29, C 30 |
|
| C 29 | 117.700 | 121.480 | CH | 5.110 | C 31, C 32, C 28 |
|
| C 30 | 135.600 | 135.680 | C | |||
| C 31 | 18.100 | 18.100 | CH3 | 1.660 | C 32, C 29, C 30 |
|
| C 32 | 25.800 | 25.840 | CH3 | 1.710 | ||
| O 1 | OH | 3.690 | C 2, C 1 |
The Molecular Connectivity Diagram MCD was automatically generated (Fig. 1) by Structure Elucidator. The atomic properties defined by the authors were used with the only exception of setting carbons at 86.1, 88.2 and 94.3 as of indefinite hybridization (black atoms) as they may really be of any kind and there is no definite proof or preference for any.
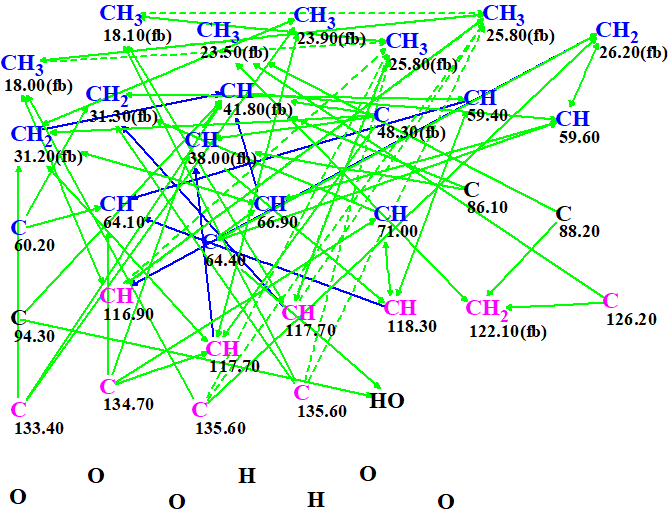
Figure 1
We also see that the MCD contains ambiguous correlations (dotted lines). These appear because of the presence of the three overlapping 13C shifts and are shown better in Figure 2. Even though these create an uncertainty in the data entered to the program, they are by no means a show stopper. The only real downside is that the elucidation will take longer as all the combination of options will need to be explored.
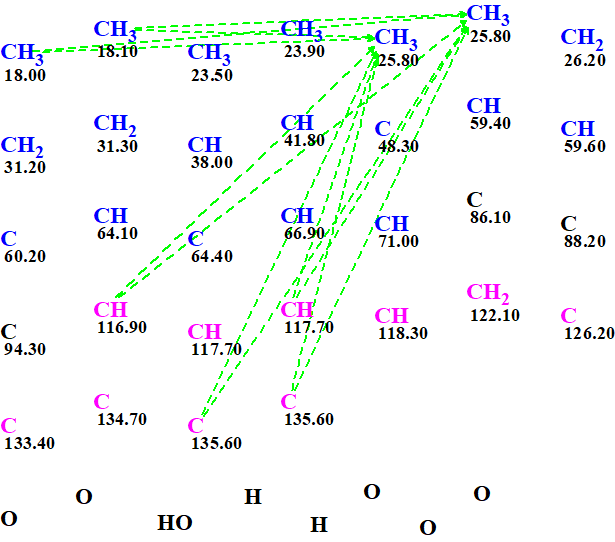
Figure 2
Strict generation was initiated together with 13C chemical shift prediction using the incremental method. The were 344,063 structures generated in 14 minutes and then 1283 passed the filter: k = 344063→1429→1283, tg = 14 m. The structures were then ranked according to the mean deviation between the predicted using HOSE codes and Neural Networks 13C chemical shifts and the experimental ones (dA and dN respectively). The results are summarized in Figure 3.
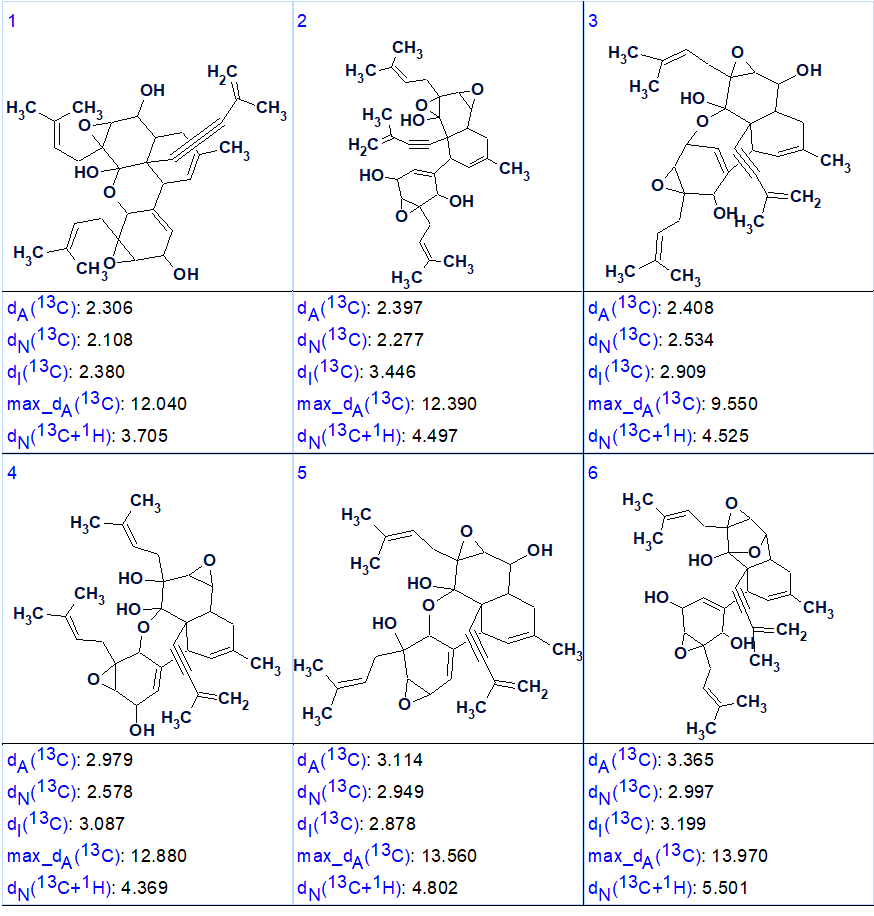
Figure 3. The first 6 structures of the ranked output file
The structure ranked as the first in figure 3 is the correct one, and is identical to 1. It is also worth noting that only the first 11 structures contained a triple bond, the other 1272 did not. This is enough to rule them out as there will probably be clear evidence of the triple bond by other methods (e.g., IR spectroscopy).
It is also worth noticing that the first 3 or 4 structures in Fig 3 have mean deviations that are very close to each other. Even though structure 1 has the lowest deviations with all three methods that were used to calculate it some further proof may be needed in order to reduce to increase the confidence. This can either be achieved by recording additional NMR spectra like 1,1 ADEQUATE of LR-HSQMBC or by calculating the chemical shifts using DFT methods [4,5]
The correct structure together with the found 13C assignments is shown in Figure 4.
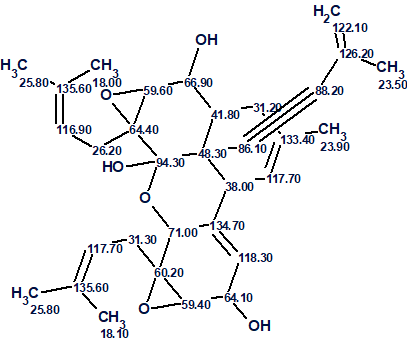
Figure 4: The elucidated structure of Dimericbiscognienyne A with the 13C assignments.
References
- H. Zhao, G.-D. Chen, J. Zou, R.-R. He, S.-Y. Qin, D. Hu, G.-Q. Li, L.-D. Guo, X.-S. Yao, H. Gao. (2017). Dimericbiscognienyne A: A Meroterpenoid Dimer from Biscogniauxia sp. with New Skeleton and Its Activity, Org. Lett., 19: 38−41.
- Elyashberg, M. E.; Williams, A. J.; Blinov, K. A. Contemporary Computer-Assisted Approaches to Molecular Structure Elucidation; RSC, Cambridge, 2012.
- M. E. Elyashberg, A. J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data. The Art of Solving Problems. Springer.
- Buevich, A. V.; Elyashberg, M. E. (2016). Synergistic combination of CASE algorithms and DFT chemical shift predictions: a powerful approach for structure elucidation, verification and revision. J. Nat. Prod., 79(12): 3105–3116.
- Buevich, A. V.; Elyashberg, M. E. (2018). Towards unbiased and more versatile NMR-based structure elucidation: A powerful combination of CASE algorithms and DFT calculations. Magn. Reson. Chem., 56(6): 493–504. DOI: 10.1002/mrc.4645


