September 1, 2017
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Emodacidamide A
Anthraquinones (also known as anthraquinonoids) are a class of naturally occurring phenolic compounds based on the 9,10-anthraquinone skeleton. They are usually found in higher plants and fungi. Fungal members of the genera Aspergillus, Pyrenochaeta, and Pestalotiopsis are known to contain many antraquinonoids. Anthraquinones are widely used as anticancer, antimalarial, and laxative agents due to their wide biological activity. Anthraquinone—amino acid conjugates have shown great potential with radiosensitizing activities in H460 human lung cancer cells in vitro. However such species have never been isolated from natural sources.
It is well known that many bioactive natural products can be found in marine-derived fungi. During screenings for new secondary metabolites from fungi originating from the South China Sea, Luo and co-workers [1] isolated 10 anthraquinone compounds, including eight new anthraquinones representing rhein−amino acid conjugates, regarded as emodacidamides A−H. Structures of these compounds were elucidated using NMR spectroscopy and mass spectrometry. The NMR data utilized for the structure elucidation of emodacidamide A (1) in [1] were used by us to challenge the ACD/Structure Elucidator.
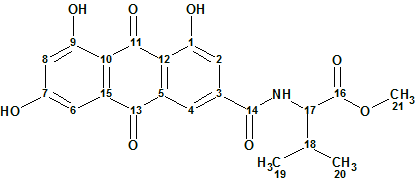
1
Compound 1 was obtained as a yellow powder, and its molecular formula found to be C21H19NO8 based on positive ion high-resolution ESI-MS, which showed a peak corresponding to the protonated molecule at m/z 414.1173 ([M + H]+). Also a sodium adduct ion peak at m/z 436.0990 ([M + Na]+) was seen. These indicated 13 degrees of unsaturation.
The NMR spectroscopic data from article [1] (13C and 1H chemical shifts with 1H multiplicities) and key HMBC correlations are shown in Table 1.
Table 1. Emodacidamide A. NMR spectroscopic data.
| C/X Label |
δC | δC calc | XHn | δH | M(J) | H to C HMBC |
| C 1 | 161 | 165.07 | C | |||
| C 2 | 123 | 120.98 | CH | 7.79 | d(1,6) | C 1, C 4, C 12, C 14 |
| C 3 | 140.5 | 136.2 | C | |||
| C 4 | 118.1 | 118.35 | CH | 8.11 | d(1,6) | C 2, C 12, C 13, C 14 |
| C 5 | 133.4 | 133.34 | C | |||
| C 6 | 109 | 107.69 | CH | 7.17 | d(2.2) | C 7, C 8, C 10, C 13 |
| C 7 | 165.9 | 164.82 | C | |||
| C 8 | 108.1 | 108.1 | CH | 6.63 | d(2.2) | C 6, C 7, C 10 |
| C 9 | 164.7 | 164.32 | C | |||
| C 10 | 109.4 | 110.04 | C | |||
| C 11 | 189.5 | 190.17 | C | |||
| C 12 | 117.5 | 119.88 | C | |||
| C 13 | 181.1 | 183.49 | C | |||
| C 14 | 165 | 165.39 | C | |||
| C 15 | 135.2 | 134.73 | C | |||
| C 16 | 171.9 | 172.55 | C | |||
| C 17 | 58.9 | 57.4 | CH | 4.32 | t(7.6) | C 14, C 16, C 18 |
| C 18 | 29.5 | 31.84 | CH | 2.21 | u | C 17, C 19, C 20 |
| C 19 | 19.1 | 17.9 | CH3 | 0.99 | d(6.8) | C 17, C 18 |
| C 20 | 19.1 | 18.9 | CH3 | 0.94 | d(6.8) | C 17, C 18 |
| C 21 | 51.8 | 52.1 | CH3 | 3.67 | s | C 16 |
| N 1 | NH | 9.09 | d(8.0) | C 14, C 16 |
||
| O 1 | OH | 12.09 | s | C 1, C 2, C 12 |
||
| O 2 | OH | 11.51 | s | |||
| O 3 | OH | 12.01 | s | C 8, C 9, C 10 |
The Molecular Connectivity Diagram (MCD) automatically produced by the program is shown in Figure 1.
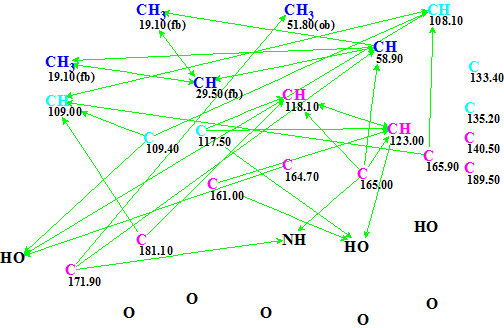
Figure 1. Emodacidamide A. Initial molecular connectivity diagram.
MCD overview. Visual analysis of the MCD shows some peculiarities:
- Six carbon atoms distinguished by blue color were of undefined hybridization not sp (sp2 or sp3)
- There is no connectivity information (HMBC peaks) for four quaternary carbon atoms (C 133.4, C 135.2, C 140.5 and C 189.5)
- More than a half of the carbon atoms (12 of 21) are quaternary
- The molecular formula of the compound C21H19NO8 is severely hydrogen-deficient with the ratio of skeletal atoms (C+N+O) to nonequivalent hydrogen atoms, R, equal to 2.3 (30/13). It is known that problems with R more than about 1.5 are becoming difficult for CASE so at 2.3 this is expected to be a very difficult one
Taking these into account some additional constraints should be introduced in order to obtain the solution to the problem in a reasonable amount of time.
In this respect:
- An sp2 hybridization was set for atoms C 133.4 and 135.2 (The only case for these to be sp3 is if they are connected to two heteroatoms, which is highly unlikely)
- Label “ob” was assigned to carbons C 161–C 189.5 to force these atoms to have at least one neighboring heteroatom.
- Multiplicities of 1H signals shown in Table 1 were used to specify the numbers of hydrogen atoms in the environment of a given hydrogenated skeletal atom.
The edited MCD is shown in Figure 2.
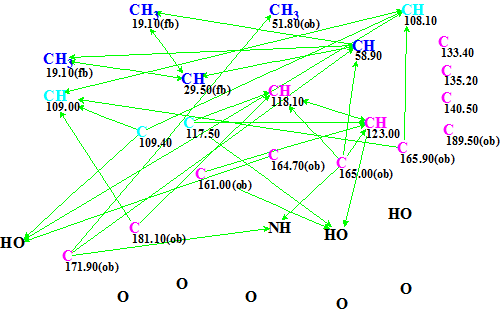
Figure 2. Emodacidamide A. Edited MCD.
Structure generation followed by 13C chemical shift prediction by incremental approach [2] and spectral filtering (structures for which average deviations exceeded 5 ppm were rejected) was initiated from the edited MCD. The generation was completed with the following results: k = 227,952 → 11 → 3, tg = 2 h 48 min. The long structure generation time is explained by the spectrum-structural information peculiarities and the fact that only minimal edits of the initial MCD were made.
13C chemical shift prediction was carried out for the structures using also two other methods implemented into ACD/Structure Elucidator—HOSE code and neural networks based. The output file ranked in increasing order of average deviation values is shown in Figure 3.
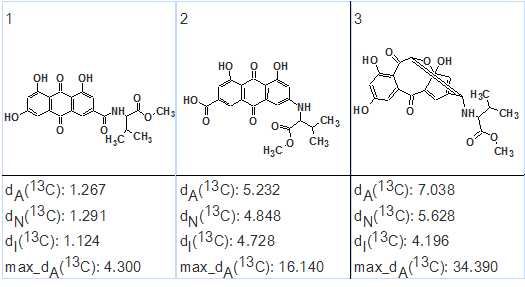
Figure 3. Emodacidamide A. The output file ranked in increasing order of average deviation values.
Figure 3 shows that the first ranked structure is the same as the structure of emodacidamide A determined by the authors of [1], while the difference between the average deviations of the first structure and the other ones is very large. This large difference serves as a reliable confirmation of the solution accuracy. The elucidated structure with 13C chemical shift assignments is shown below.
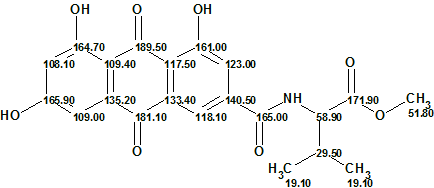
From a methodological point of view it was interesting to see how the time of structure generation will be reduced if some new additional information is added. For this reason double bonds were drawn manually from C 181.1 and 189.5 to oxygen atoms, defining the two carbonyls as shown in Figure 4. These two “axioms” look rather straightforward in the current case.
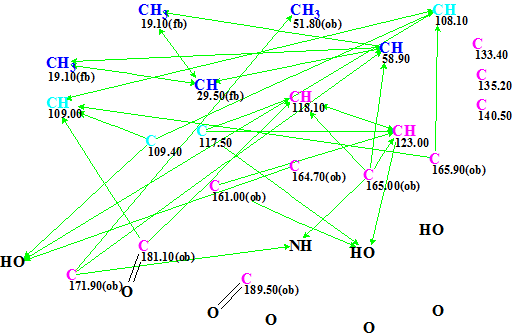
Figure 4. Emodacidamide A. MCD containing two carbonyls drawn by hand.
Results of the second structure generation run: k=1260 → 8 → 3, tg = 1 m 20 s. We see that adding just two evident carbonyl bonds reduced the generation time by 125 times, while the output file was the same. When an oxygen atom was manually connected with CH3 51.8 (the presence of OCH3 is evident) in the MCD shown in Figure 4, the generation time further reduced down to 40 s, that is by 250(!) times.
This example demonstrates how introducing additional structural information based on NMR characteristic chemical shifts and common spectroscopic sense reduces the generation time drastically without affecting the validity of the final result.
References
- M. Luo, Z. Cui, H. Huang, X. Song, A. Sun, Y. Dang, L. Lu, J. Ju. (2017). Amino Acid Conjugated Anthraquinones from the Marine-Derived Fungus Penicillium sp. SCSIO sof101. J. Nat. Prod. 80: 1668−1673.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.


