January 1, 2016
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Flueggether A
Securinega alkaloids are an interesting class of natural products discovered only from plants of a limited number of genera, such as Securinega and Flueggea of the Euphorbiaceae family. This structure family, with the rigid tetracyclic backbone incorporating a butenolide moiety, has attracted much attention from both natural products and synthetic chemists. Securinega alkaloids reemerged as a hot research topic in the chemical community in this century.
The Flueggea genus is a small tribe with about 12 species distributed all over the world. F. virosa (Roxb. ex Willd.) Voigt is the star plant among the four native Chinese species due to its diverse secondary metabolites. H. Zhang et al [1] observed interesting signals from very minor constituents from F. virosa in their MS analysis. A further fractionation of the fractions showing the interesting MS peaks returned two extra new alkaloids, one of which, Flueggether A, (1) is the first example of Securinega alkaloid oligomers which features an ether bond linkage.

1
The structure of 1 was characterized on the basis of comprehensive spectroscopic data analysis, and its absolute configuration was established by a single-crystal X-ray diffraction experiment and comparison of the electronic circular dichroism (ECD) spectrum with the calculated one.
Flueggether A was obtained as colorless crystals and displayed IR absorption bands at 1753 and 1651 cm-1, which can correspond to conjugated γ-lactone(s). The molecular formula of C25H30N2O5 was assigned to 1 on the basis of 13C NMR data and (+)-HRESIMS analysis at m/z 439.2228 ([M +H]+, Δmmu -0.5), consistent with 12 degrees of unsaturation.
The spectroscopic NMR data used by authors [1] for elucidating the structure of Flueggether A are presented in Table 1.
Table 1. Spectroscopic NMR data for Flueggether A.
| C Label | δC | δC calc | XHn | δH | COSY | C HMBC |
| C 1 | 63.7 | 61.31 | CH | 2.23 | 1.66 | C 9 |
| C 2 | 26 | 25.27 | CH2 | 1.66 | 1.87, 2.23 | |
| C 2 | 26 | 25.27 | CH2 | 1.35 | ||
| C 3 | 24.8 | 21.4 | CH2 | 1.28 | ||
| C 3 | 24.8 | 21.4 | CH2 | 1.87 | 1.58, 1.66 | |
| C 4 | 26.9 | 25.59 | CH2 | 1.54 | ||
| C 4 | 26.9 | 25.59 | CH2 | 1.58 | 1.87, 2.81 | |
| C 5 | 52.7 | 50.41 | CH2 | 2.67 | ||
| C 5 | 52.7 | 50.41 | CH2 | 2.81 | 1.58 | C 1 |
| C 6 | 56.2 | 63.04 | CH | 2.94 | 2.90, 3.87 | C 1, C 5 |
| C 7 | 72.3 | 72.6 | CH | 3.87 | 2.69, 2.94 | C 25 |
| C 8 | 34 | 35.68 | CH2 | 1.2 | ||
| C 8 | 34 | 35.68 | CH2 | 2.69 | 3.87 | C 9, C 12 |
| C 9 | 84.7 | 83.87 | C | |||
| C 10 | 174.2 | 172.77 | C | |||
| C 11 | 109.1 | 116.47 | CH | 5.62 | C 9, C 10, C 12 | |
| C 12 | 176 | 171.01 | C | |||
| C 13 | 23.7 | 25.96 | CH2 | 2.77 | ||
| C 13 | 23.7 | 25.96 | CH2 | 2.9 | 2.94 | C 9, C 12 |
| C 14 | 66 | 70.86 | CH | 3.07 | 1.92 | |
| C 15 | 29.1 | 27.26 | CH2 | 1.74 | ||
| C 15 | 29.1 | 27.26 | CH2 | 1.92 | 1.96, 3.07 | |
| C 16 | 26.8 | 23.82 | CH2 | 1.96 | 1.92, 3.31 | |
| C 17 | 57.4 | 52.6 | CH2 | 3.31 | 1.96 | C 14, C 18 |
| C 17 | 57.4 | 52.6 | CH2 | 2.64 | ||
| C 18 | 64.6 | 61.58 | CH | 3.01 | 2.29, 3.77 | |
| C 19 | 30.2 | 40.71 | CH2 | 1.74 | ||
| C 19 | 30.2 | 40.71 | CH2 | 2.29 | 3.01 | C 14, C 20, C 22 |
| C 20 | 91.5 | 90.2 | C | |||
| C 21 | 172.8 | 172.36 | C | |||
| C 22 | 111.3 | 114.17 | CH | 5.64 | C 20, C 21, C 22 | |
| C 23 | 172.6 | 173.01 | C | |||
| C 24 | 29.4 | 33.11 | CH2 | 2.8 | 3.77 | C 22 |
| C 25 | 74.3 | 78.01 | CH | 3.77 | 2.80, 3.01 |
These data were input into ACD/Structure Elucidator Suite, and the Molecular Connectivity Diagram (MCD) was created by the program (Figure 1).
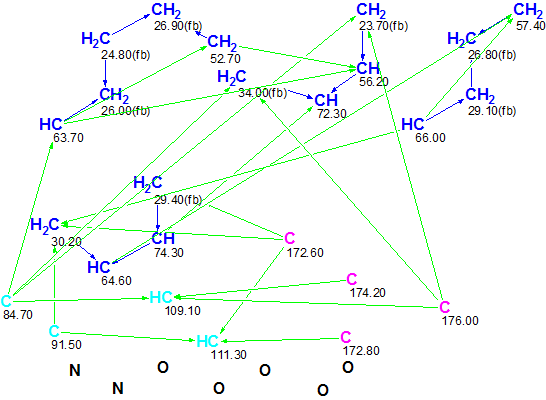
Figure 1. Molecular Connectivity Diagram (MCD) for Flueggether A. HMBC connectivities are marked by green arrows, COSY connectivities by blue arrows.
MCD overview. The following structural and spectroscopic peculiarities can be noted from the MCD:
- Four carbon atoms with chemical shifts between 84.70 and 111.3 ppm (colored in light blue color) were accepted by the program as those having hybridization “not sp” (sp2 or sp3). The program took into account the possibility of forming either C=C or O-(C<)-O fragments by these atoms.
- Four atoms with chemical shifts between 172.60 and 176.00 ppm (colored in violet color) were accepted by the program as having hybridization sp2. At the same time, no descriptors indicating the possibility of neighboring with heteroatoms (fb – forbidden, ob – obligatory) were assigned to these atoms. It is worthy to note that the mentioned chemical shifts are typical for ester and amide groups, as well as for O-C=C and C=N and there is the temptation to mark these atoms as ones conned to heteroatoms, but it is evident that the right solution could be lost by doing this. Strictly speaking, when structure elucidation is performed, ALL possibilities should be checked, which may take a very long time if this was done manually. When Structure Elucidator Suite is used, this work can be done by the program.
- All remaining carbon atoms were marked by the program as sp3-hybridized (colored in blue color), while only part of them received descriptor fb (both 13C and 1H chemical shifts indicate that neighboring with heteroatom is impossible).
No further edits of MCD were made. Checking the MCD for 2D NMR data consistency showed that all COSY and HMBC connectivities are of standard lengths [2], therefore Strict Structure Generation combined with 13C chemical shift prediction (using the incremental method) and spectral and structural filtering was initiated. In so doing, generated structures for which the average deviation of predicted chemical shifts exceeded 5 ppm were rejected by the program.
The results were: k = 21,959 → 3, tg = 13m 40s, meaning that only three (of almost 22 thousand) structures survived the filtering, while the structure generation, 13C chemical shift prediction and structure filtering process took almost 14 minutes. According to the general methodology, 13C chemical shift prediction using HOSE code based and neural networks approaches was performed for the structures included into the output file. The output file, ranked in ascending order of the average deviation value, is presented in Figure 2.
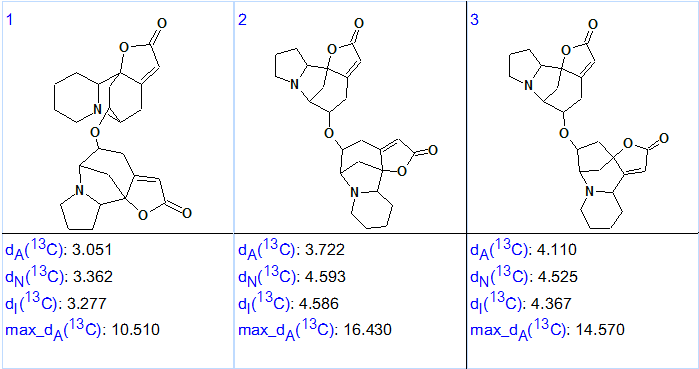
Figure 2. Ranked output structural file for Flueggether A. dA is an average deviation for HOSE code based approach, dI for the incremental method, dN for the neural networks approach.
Figure 2 shows that the first ranked structure coincides with structure 1 which was confirmed by single-crystal X-ray diffraction experiment in [1]. The elucidated structure with automatically assigned 13C chemical shifts is shown below:
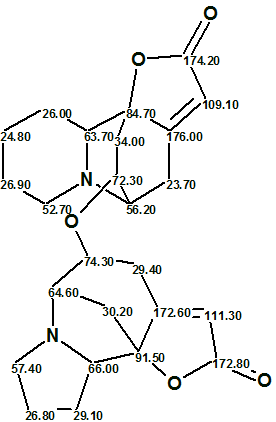
We see that 13C chemical shifts 172.60 and 176.00 pertain to carbon atoms which have no connections to heteroatoms. A structural search against ACD/CNMNR Database (425,000 entries) has shown that these shift values are characteristic for conjugated γ-lactones.
In conclusion, ACD/Structure Elucidator Suite was able to determine the correct structure of a new complex natural product Flueggether A (published in Organic Letters in December 2015) in a fully automatic process, in ~15 min.
References
- H. Zhang, K.-K. Zhu, Y.-S. Han, C. Luo, M. A. Wainberg, J.-M. Yue. (2015). Flueggether A and Virosinine A, Anti-HIV Alkaloids from Flueggea virosa. Org. Lett., 17(24):6274–6277.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.


