June 1, 2013
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Geralcin A
Hydrazines and hydrazides are structural motifs often found in synthetic therapeutics that are in clinical use. An important example is iproniazid, a hydrazine monoamine oxidase inhibitor, used as an antidepressant. In contrast, natural hydrazide compounds are notably scarce, and only four such structures have been reported to date.
Le Goff et al [1] reported the structural characterization of two novel alkylhydrazides produced by the bacterial strain Streptomyces sp. LMA-545. The structures were elucidated using both 1D and 2D 1H and 13C NMR spectroscopic analysis and high-resolution mass spectrometry. 1H–15N NMR experiments were required for full structural elucidation. Here we investigate the structure elucidation of Geralcin A (1), one of the three isolated novel compounds. This compound consists of a novel natural scaffold that connects an alkylhydrazide to an α,β-unsaturated γ-lactone.
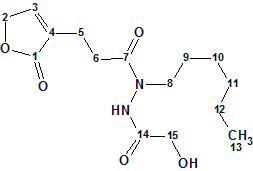
1
Compound 1 was obtained as yellowish oil. The HRESIMS analysis gave the molecular formula C15H24N2O5: m/z [M + Na]+ 335.1575 (calculated for C15H24N2O5Na, 335.1583). IR absorption bands observed at 3430 and 3275 cm-1 indicate the respective presence of NH and OH functionalities. An absorption band at 1741 cm-1 is present, and characteristic for a carbonyl group. The NMR data acquired for the structure elucidation are listed in Table 1.
Table 1. Geralcin A. Spectroscopic NMR data.
| Label | δX | δX calc | XHn | δH | M(J) | COSY | C HMBC | N HMBC |
| C 1 | 174.4 | 173.71 | C | |||||
| C 2 | 70.8 | 70.08 | CH2 | 4.85 | u | 7.48 | C 1, C3, C 4 | |
| C 3 | 147 | 146 | CH | 7.48 | u | 4.85 | C 2, C 5, C 1, C 4 | |
| C 4 | 132.3 | 132.86 | C | |||||
| C 5 | 20.7 | 20.18 | CH2 | 2.61 | u | 2.47 | C 7, C 4, C 6, C 3 | |
| C 6 | 30.5 | 30.7 | CH2 | 2.47 | u | 2.61 | C 5, C 4, C 7 | |
| C 7 | 173.2 | 171.63 | C | |||||
| C 8 | 47.4 | 49.09 | CH2 | 3.58 | u | C 9, C 7 | N 2 | |
| C 9 | 27.2 | 26.21 | CH2 | 1.52 | u | C 8, C 11, C 10 | N 1 | |
| C 10 | 26.6 | 26.92 | CH2 | 1.28 | u | |||
| C 11 | 31.8 | 31.43 | CH2 | 1.28 | u | |||
| C 12 | 22.7 | 22.56 | CH2 | 1.28 | u | |||
| C 13 | 13.9 | 13.95 | CH3 | 0.86 | t(6.9) | C 12, C 11 | ||
| C 14 | 171.8 | 171.18 | C | |||||
| C 15 | 62 | 62.18 | CH2 | 4.14 | d(5.9) | 5.79 | C 14 | |
| N 1 | 133.5 | 139.53 | N | |||||
| N 2 | 137 | 129.66 | NH | 10.44 | s | C 15, C 14, C 7 | N 1 | |
| O 1 | 100* | OH | 5.79 | t(5.9) | 4.14 | C 15, C 14 |
* Fictitious 17O chemical shift.
To simplify the structure elucidation of Geralcin A, the authors [1] also included the 1H–15N HMNC spectrum. The 1H–15N HMBC data revealed two nitrogen atoms at δN 133.5 (N-1, N) and 137.0 (N-2, NH). Table 1 shows that the proton d10.44 attached to a nitrogen atom produces correlations to carbons C 15, C 14 and C 7, and to N 1. Three correlations to nitrogen atoms are presented in the column N HMBC. The correlations of H-N (δH 10.44, s, NH) to N-1 (δN 133.5, N) and (δH 3.58, m) to N-1 (δN 133.5, N) suggests that a N-N bond corresponding to a hydrazide group is present. The spectrum-structural information contained in the Table 1 is presented graphically on Molecular Connectivity Diagram (MCD, Figure 1).
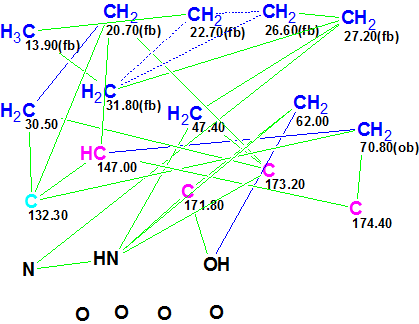
Figure 1. Geralcin A. Molecular Connectivity Diagram.
MCD overview. The authors [1] state that there are three carbonyl atoms at δC 174.4 (C-1), 173.2 (C-7), and 171.8 (C-14), but properties of these atoms were not edited on the MCD (C=O bonds were not drawn by hand) because the same 13C chemical shifts are also characteristic of the sp2 hybridized carbons connected to an oxygen by a ordinary bond (C=C-O-). Ambiguous COSY correlations (dotted lines) are due to three overlapping 1H signals at 1.28 ppm related to protons attached to atoms C 10, C 11 and C 12. The numbers of hydrogens attached to neighbor skeletal atoms were set on the MCD in accordance with multiplicities listed in Table 1 (column M(J)). Note that multiplicities of 1H signals produced by NH and OH groups were also taken into account.
First run. For the first run it was assumed that there were no hints to the existence of N-N chemical bond in a molecule. When chemical bonds between heteroatoms were forbidden, MCD checking by the software revealed contradictions in 2D NMR data. Fuzzy Structure Generation was run with “Determine Options Automatically” selected. The results were: k=1, (i.e., one structure, 2, was generated) tg=1.5 s, 2 from 26 connectivities have been extended and 325 from 325 connectivity combinations were used. However all deviations calculated for the structure 2 (red arrows show nonstandard connectivities) were too large (d~5.5 ppm) for it to be considered correct.
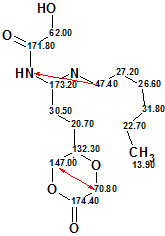
2
The results obtained were thus considered a hint for trying structure generation with options allowing chemical bonds between heteroatoms of the same
kind.
Second run. The corresponding checkboxes were selected and the structure generation gave the following results: k=1, tg=0.001 s, all deviations are less than 1 ppm. Structure 1a was obtained, which coincides with the structure of Geralcin A.
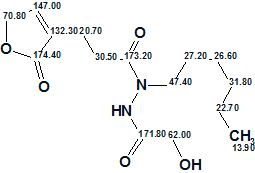
1a
Therefore a correct solution was found instantly and almost automatically using the spectral data presented in Table 1.
Third run. For completeness, it is interesting to know if Structure Elucidator is capable of solving this problem without any application
of H-N HMBC data. The structure generation was repeated with the options shown in Figure 2.

Figure 2. Options of the structure generation with H-N NMBC data switched off.
The following results were obtained: k=49→31→9, tg=0.03 s, and the first six structures of the ranked output file are presented
in Figure 3.
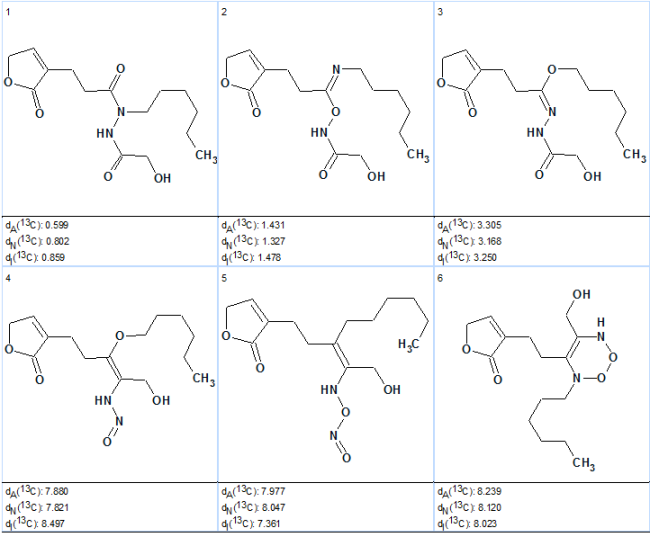
Figure 3. First six structures of the ranked output file obtained without employing H-N HMBC data.
The best structure, #1, which is characterized with very small deviations is identical to the structure of Geralcin A. Deviations of 13C chemical shifts predicted for structure #2 are greater, and though their values are also acceptable, the priority of structure #1 is evident. Note that structure #2 which is characterized with small deviations (1.3–1.5 ppm) contains only two carbonyl groups (not three as structure #1) , and its generation became possible due to the refusal to draw carbonyl bonds at carbons C 174.4, C 173.2 , and C 171.8.
The third run demonstrates that the utilization of soft constraints during MCD analysis and editing allows one to more completely consider all structural hypotheses. Overall this example shows that a compound which contains a novel natural scaffold was identified with the aids of Structure Elucidator instantly and without acquiring 1H–15N HMBC data.
[1] Isolation and Characterization of α,β-Unsaturated γ-Lactono-Hydrazides from Streptomyces sp. G. Le Goff, M.-T. Martin, C. Servy, S. Cortial, P. Lopes, A. Bialecki, J. Smadja, J. Ouazzani.J. Nat. Prod., 75(5):915–919, 2012.

