June 1, 2014
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Gymnopalyne A
Novel antibiotics with unusual scaffolds and new modes of actions are urgently needed, since the resistance against antibiotics in use is constantly increasing. Most of the known antibiotics are derived from fungal or microbial cultures. In the course of search for novel antibiotics, Thongbai et al [1] have focused on tropical basidiomycetes from Asia. In an antimicrobial screen, extracts prepared from submerged cultures of this strain showed prominent activity. The research led to the isolation of unprecedented antimicrobial metabolite Gymnopalynes A (1).
1
Though the molecule is small and relatively simple, elucidation of its structure with the aids of CASE approach is not straightforward. With this in mind we used this problem to illustrate some nuances associated with utilization of ACD/Structure Elucidation.
Gymnopalyne A (1) was obtained as a colorless oil. The presence of a chlorine atom was indicated by its characteristic isotopic pattern in the HRESIMS, which also provided the molecular formula of structure 1 as C12H7O2Cl, implying 9 degrees of unsaturation. Absorption bands observed in the IR spectrum at 1720 and 1600 cm-1 suggest the presence of carbonyl and a benzene ring respectively. As the number of skeletal atoms is twice the number of hydrogens contained in the molecular formula, the problem of elucidation can be considered as challenging in accordance to Crews’ rule. To elucidate the structure of Gymnopalyne A, the spectroscopic NMR data presented in Table 1 were used.
Table 1: Gymnopalyne A. Spectroscopic NMR data.
| Label | δC | δC calc | CHn | δH | M(J) | COSY | C HMBC |
| C 1 | 163.1 | 161.07 | C | ||||
| C 2 | 138.2 | 134.78 | C | ||||
| C 3 | 113.8 | 112.80 | CH | 7.02 | u | C 4, C 2, C 5, C 9, C 10 | |
| C 4 | 137.8 | 136.06 | C | ||||
| C 5 | 127.6 | 126.00 | CH | 7.6 | u | 7.82 | C 3 |
| C 6 | 136.5 | 134.9 | CH | 7.82 | u | 7.6 | |
| C 7 | 130.8 | 129.8 | CH | 7.62 | u | 8.24 | |
| C 8 | 130.4 | 129.35 | CH | 8.24 | u | 7.62 | C 1, C 4 |
| C 9 | 122.7 | 121.08 | C | ||||
| C 10 | 79.1 | 75.98 | C | ||||
| C 11 | 89.8 | 91.17 | C | ||||
| C 12 | 30.6 | 31.34 | CH2 | 4.55 | u | C 2, C 3, C 11, C 10 |
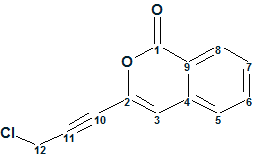
As the original work presented no evidence of a CºC triple bond [1], the Molecular Connectivity Diagram (MCD, Figure 1) was first created with the option “Allow sp Carbons” being deselected.
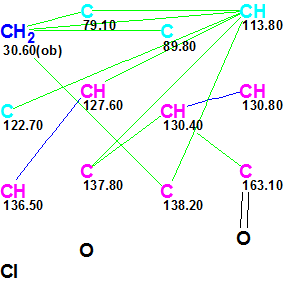
Figure 1. The first Gymnopalyne A. Molecular Connectivity Diagram (MCD), where triple bonds were not expected in the structure.
MCD overview. Four carbon atoms (C 79.10, C 89.80, CH 113.80, and C 122.70) are colored in light blue color indicating that hybridization of these atoms is sp3 or sp2. To decrease the number of conceivable structures during structure generation, two obvious constraints were introduced into the MCD: atom CH2 (30.60, 4.55) was marked with the label “ob” (i.e., a neighboring heteroatom, most probably a chlorine in this case, is obligatory) and the carbonyl group was drawn by hand at the carbon C 163.10. The software did not reveal the presence of any contradictions (using the “Check MCD” function), so strict structure generation accompanied with 13C chemical shift prediction (with d13C=5 ppm as a threshold) was initiated. Generation was completed in one second with empty structural file: all generated structures were rejected due to huge values of average deviations calculated.
Based on this result, it was proposed that at least one latent (implicit) nonstandard correlation is present in the HMBC data, and Fuzzy Structure Generation was performed with the options: m=1-20, a=16, “Stop generation when structures generated”. 13C chemical shift calculation was not switched on during the generation. Results: k=987316→8→4, tg =10 m and the structures generated are shown in Fig. 2.
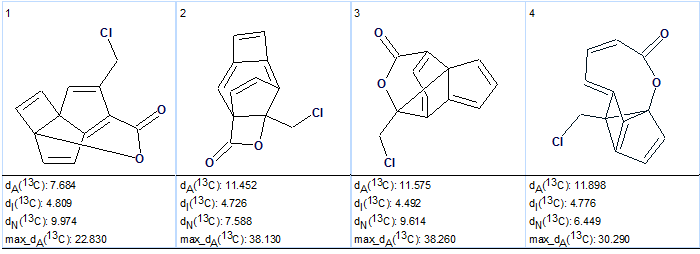
Figure 2. The ranked output structural file for Gymnopalyne A. obtained by Fuzzy Structure Generation, given sp carbon atom hybridization is forbidden.
The figure convincingly demonstrates that the solution is wrong (very large average deviations, while structures are too exotic). Therefore it was suggested that possibility of the presence of triple bonds in the molecule should be investigated.
The Molecular Connectivity Diagram was created anew with the option “Allow sp Carbons” being selected. The slightly edited MCD is presented in Figure 3.
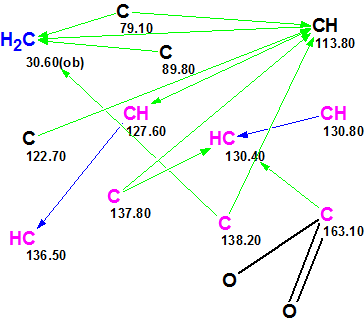
Figure 3. The slightly edited Molecular Connectivity Diagram for Gymnopalyne A.
In this MCD, four carbon atoms (C 79.10, C 89.80, CH 113.80 and C 122.70) are now colored in black, which means that hybridization states of these atoms failed to be set automatically because all hybridization types (sp3, sp2 and sp) are allowed for them if the presence of triple bonds is permitted. Thus all admissible hybridizations of these atoms will be tried during structure generation. To decrease the number of conceivable structures, the evident ester group was drawn by hand at the carbon C 163.10. MCD checking again did not reveal the presence of any contradictions and, as above, the strict structure generation combined with 13C chemical shift calculation was initiated, which also quickly produced empty structural file. Therefore Fuzzy Structure Generation was run with the following options: m=1-20, a=16, “Stop generation when structures generated”. Results: k=17967→4→1, tg=24 s, where 2 from 13 connectivities were extended during generation, and 78 of 78 possible connectivity combinations were used during generation. The single structure coincided with the proposed structure of Gymnopalyne A and its correctness was confirmed by small values of average deviations (dA13C =1.7 ppm). Assignments of 13C chemical shifts to carbon atoms along with nonstandard HMBC connectivities (their lengths are of three and four skeletal bonds) are shown in red on structure 1a:
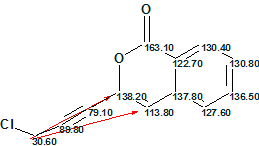
1a
It was interesting to learn if the presence of these nonstandard correlations could be suggested from visual analysis of HMBC pattern. Figure 4 shows part of the Gymnopalyne A HMBC experimental spectrum.
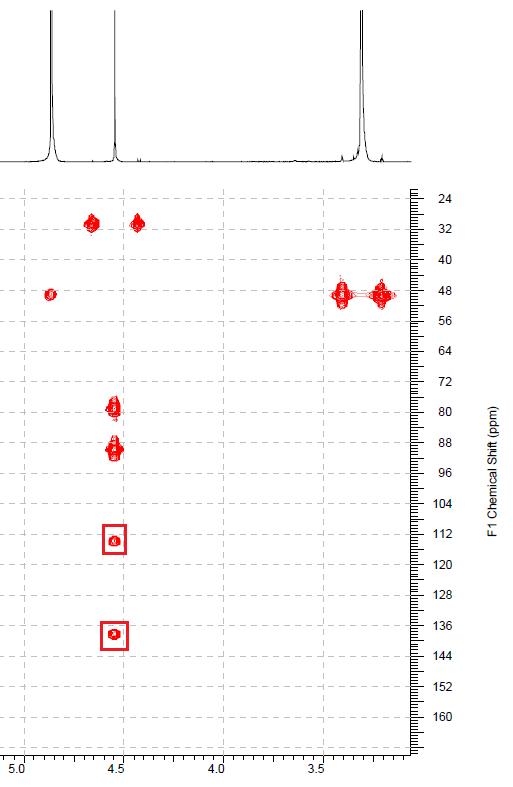
Figure 4. Fragment of the HMBC experimental spectrum of Gymnopalyne A. Peaks corresponding to the nonstandard connectivities are marked by squares
Figure 4 shows that intensities of two peaks corresponding to nonstandard correlations (in squares) are comparable with those belonging to other peaks, which gives no ground for suspecting the peaks 4.55 to 138.2 and 4.55 to 113.8 ppm to be of nonstandard length. At the same time, the software did not find discrepancies in the data to indicate that non-standard correlations might be present.
However, after failing to generate a reasonable structure in either standard mode or with Fuzzy Structure Generation, the software quickly generated the correct structure (and only that structure) when nonstandard connectivities and triple-bond hybridization were considered. Since this is a logical progression in the steps taken to solve an elucidation problem, this example demonstrates the efficiency of tackling the elucidation of unique, unprecedented structures with this ACD/Structure Elucidator Suite.
References
- Thongbai B, Surup F, Mohr K, Kuhnert E, Hyde KD, Stadler M. 2013. Gymnopalynes A and B, Chloropropynyl-isocoumarin Antibiotics from Cultures of the Basidiomycete Gymnopus sp, J. Nat. Prod., 76, 2141-2144.


