September 1, 2020
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Hypocrolide A
A particular type of sesquiterpenoids are botryanes. The have the same basic skeleton of a bivyvliv nonisoprenoid system. They have only been found in a few fungal species, including the pathogen Botrytis cinereal, a necrotrophic plant. Since they are sesquiterpene metabolites they expected, and they do show, various biological activity.
Yuan at al [1] isolated the fungus Hypocrea sp. Upon fractionation of its extract hypocrolide A (1), a mixed-biogenetic botryane derivative possessing a previously undescribed hexacycle, was isolated.
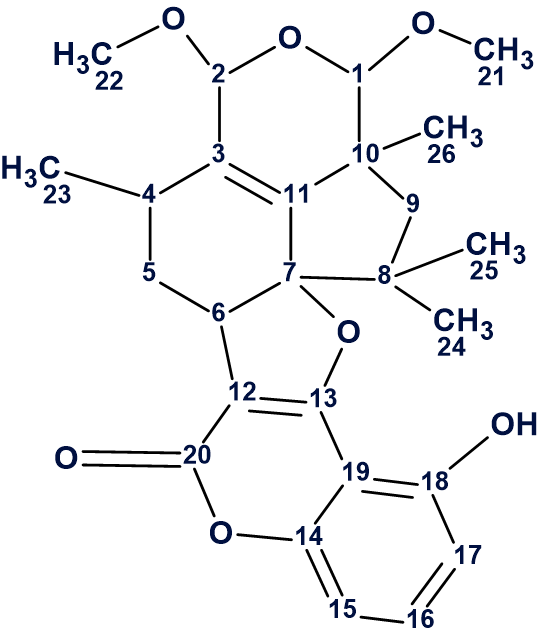
1
The proposed structure was confirmed by X-ray crystallography.
A molecular formula of C26H30O7 (12 degrees of unsaturation) was assigned to hypocrolide A (1) on the basis of HR-ESI-MS (m/z 455.2064 [M + H]+.
The 1D and 2D NMR data used for the hypocrolide A structure elucidation were used for challenging ACD/Structure Elucidator. The data used are presented in Table 1.
Table 1. NMR spectroscopic data.
| Label | δC | δC calc, HOSE | XHn | δH | M | H to C HMBC | COSY |
| C 1 | 101.900 | 105.410 | CH | 4.500 | s | C 26, C 10, C 9, C 21, C 2, C 11 | |
| C 2 | 100.400 | 95.520 | CH | 5.070 | s | C 4, C 10, C 22, C 1, C 3, C 11 |
|
| C 3 | 137.700 | 135.420 | C | ||||
| C 4 | 28.400 | 30.750 | CH | 2.380 | u | C 23, C 6, C 5, C 2, C 3 |
2.00 |
| C 5 | 35.400 | 33.170 | CH2 | 1.890 | u | C 23, C 4, C 6, C 12, C 7, C 3 |
|
| C 5 | 35.400 | 33.170 | CH2 | 2.000 | u | 2.38, 3.45 |
|
| C 6 | 33.400 | 48.380 | CH | 3.450 | u | C 4, C 5, C 8, C 12, C 7, C 11, C 20, C 13 | 2.00 |
| C 7 | 106.100 | 95.410 | C | ||||
| C 8 | 45.300 | 42.240 | C | ||||
| C 9 | 47.700 | 48.520 | CH2 | 1.810 | u | C 26, C 25, C 24, C 10, C 8, C 1, C 7, C 11 | |
| C 9 | 47.700 | 48.520 | CH2 | 1.740 | u | ||
| C 10 | 41.700 | 45.800 | C | ||||
| C 11 | 138.800 | 150.700 | C | ||||
| C 12 | 105.700 | 100.780 | C | ||||
| C 13 | 164.700 | 167.730 | C | ||||
| C 14 | 155.500 | 154.370 | C | ||||
| C 15 | 108.900 | 109.320 | CH | 6.920 | s | C 19, C 17, C 14, C 13 |
7.42 |
| C 16 | 133.500 | 130.920 | CH | 7.420 | t | C 19, C 15, C 17, C 18, C 14 | 6.76, 6.92 |
| C 17 | 110.900 | 110.950 | CH | 6.760 | s | C 19, C 15, C 18 |
7.42 |
| C 18 | 153.400 | 152.100 | C | ||||
| C 19 | 101.300 | 101.140 | C | ||||
| C 20 | 160.000 | 161.730 | C | ||||
| C 21 | 57.000 | 55.040 | CH3 | 3.560 | s | C 1 | |
| C 22 | 55.900 | 54.380 | CH3 | 3.540 | s | C 2 | |
| C 23 | 17.900 | 20.510 | CH3 | 1.170 | d | C 4, C 5, C 3 |
|
| C 24 | 27.100 | 24.160 | CH3 | 0.960 | s | C 25, C 8, C 9, C 7 |
|
| C 25 | 22.900 | 23.030 | CH3 | 1.040 | s | C 24, C 8, C 9, C 7 |
|
| C 26 | 21.300 | 19.550 | CH3 | 1.290 | s | C 10, C 9, C 1, C 11 |
|
| O 1 | OH | 7.200 | s | C 19, C 17, C 18 |
The data were entered into the program which produced the molecular connectivity diagram (MCD) shown in Figure 1.
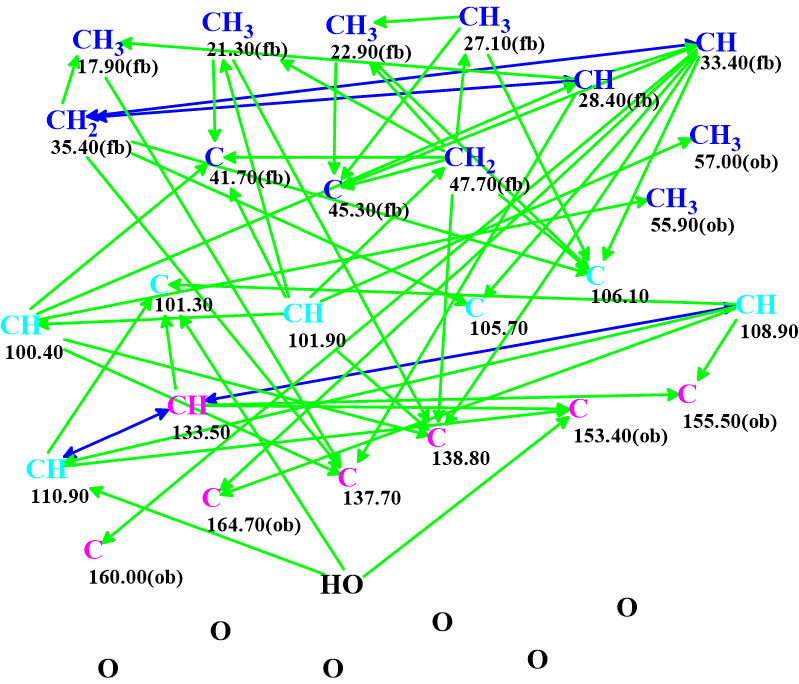
Figure 1. Molecular connectivity diagram.
The peculiarity of this MCD is the presence of seven carbon atoms marked by light blue color, which means that the atoms have been assigned ambiguous hybridization: either sp2 or sp3. They are all in the range of 100-110 ppm. For each of such carbons, both the possible hybridizations should be tried during structure generation, essentially doubling the MCD. This leads to increase of the generation time, as in fact 27 = 128 MCD with unambiguous hybridizations need to be evaluated. In the last version of ACD/Structure Elucidation, v2020.1, this difficulty was overcome by improving the algorithm of structure generation.
Checking the MCD for consistency indicated that the combined HMBC and COSY data contain a minimum two non-standard connectivities (NSCs), i.e. two connectivities that the topological distances between interacting nuclei are longer than three chemical bonds. Consequently, the Fuzzy Structure Generation [2] option was enabled initiated with the program setting the options automatically. Results: k= 1212 → (spectral filtering) → 12 → (identical structure removal) → 8, tg = 47 s, and 3 (from 55) connectivities have been extended during generation.
Then 13C chemical shift prediction was performed for all structures of the output file using three empirical methods – HOSE code based, Incremental and Neural Networks, and the structures were ranked in increasing order of average deviations dA(13C) between experimental chemical shifts and predicted ones (the deviations dA(13C) are related to the HOSE code based approach) The ranked structures are shown in Figure 2.
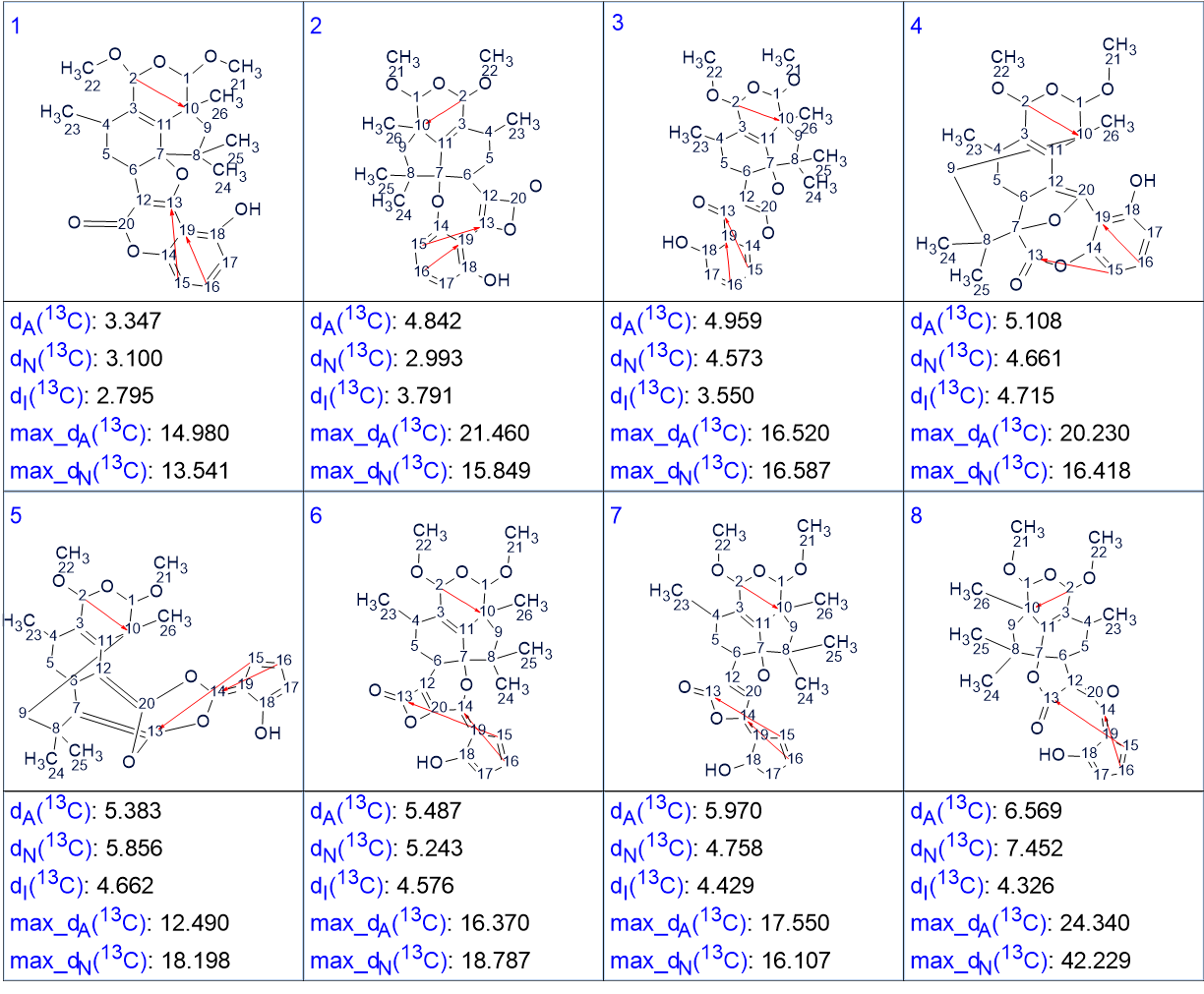
Figure 2. The ranked output structural file.
Figure 2 shows that the first ranked structure coincides with the structure of hypocrolide A suggested by the authors [1]. It contains three NSCs of three C-C bonds length (4JCH) which are marked with red arrows in the structure.
Therefore, the structure of a new natural product characterized with a new skeleton was automatically elucidated in the presence of three NSCs whose number and lengths were not known. The NSCs were automatically detected by the program during Fuzzy Structure Generation and their lengths were adjusted.
The structure of hypocrolide A together with the assigned 13C chemical shifts is shown below.
References
- Y. Yuan, Y. Feng, F. Ren, S. Niu, X. Liu, Y. Che. (2013). A Botryane Metabolite with a New Hexacyclic Skeleton from an Entomogenous Fungus Hypocrea sp. Organic Letters, 15: 6050–6053.
- M. E. Elyashberg, A. J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data. The Art of Solving Problems. Springer.


