March 28, 2023
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
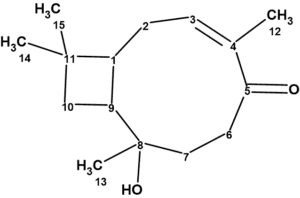
Rumphellol A
The group of Sung et. al. [1] isolated two new caryophyllene-type sesquiterpenoids, rumphellol A and B, from the Rumphella antipathies gorgorian coral off the waters of Taiwan. Their structures were elucidated using HR-ESI-MS and 1D and 2D NMR spectra (HSQC, HMBC and COSY). These compounds contain a 9-membered ring fused with a cyclobutane ring, which is rarely observed in natural products. We used ACD/Structure Elucidator to confirm the structure of rumphellol A (1) suggested by the authors [1].
1
Rumphellol A (1) was isolated as a colorless oil, and the molecular formula of this compound was determined to be C15H24O2 by high resolution electrospray ionization mass spectrometry (HRESIMS) at m/z 237.1836 (calcd. for C15H24O2 + H, 237.1849). IR absorption bands at νmax 3429 cm-1 (broad) and 1724 cm−1 revealed the presence of hydroxy and carbonyl groups.
The 13C , 1H, HSQC and HMBC data presented in Table 1 were entered into ACD/SE. The COSY data were not used in order to avoid over-constraining the problem.
Table 1. NMR spectroscopic data of rumphellol A.
| Label | δC | δCcalc (HOSE) | CHn | δH | M(H) | H to C HMBC |
| C 1 | 44.4 | 43.71 | CH | 1.83 | u | C 11, C 9, C 8 |
| C 2 | 27.6 | 30.67 | CH2 | 1.84 | u | C 1, C 9, C 3, C 4 |
| C 2 | 27.6 | 30.67 | CH2 | 2.2 | u | |
| C 3 | 128.8 | 137.48 | CH | 5.64 | u | C 12, C 2, C 5 |
| C 4 | 138.4 | 138.52 | C | |||
| C 5 | 212.7 | 203.75 | C | |||
| C 6 | 38 | 32.72 | CH2 | 2.53 | u | |
| C 6 | 38 | 32.72 | CH2 | 2.19 | u | C 7, C 8, C 4, C 5 |
| C 7 | 37.7 | 36.15 | CH2 | 2.03 | u | |
| C 7 | 37.7 | 36.15 | CH2 | 1.88 | u | C 13, C 6, C 9, C 8, C 5 |
| C 8 | 72.6 | 73.76 | C | |||
| C 9 | 44.9 | 50.6 | CH | 1.95 | u | C 2, C 10, C 1 |
| C 10 | 33.6 | 37.19 | CH2 | 1.56 | u | |
| C 10 | 33.6 | 37.19 | CH2 | 1.63 | u | C 15, C 14, C 11, C 1, C 9, C 8 |
| C 11 | 32.8 | 34.24 | C | |||
| C 12 | 19.8 | 20.96 | CH3 | 1.82 | s | C 3, C 4, C 5 |
| C 13 | 25.3 | 29.12 | CH3 | 1.02 | s | C 7, C 9, C 8 |
| C 14 | 29.6 | 30.27 | CH3 | 0.96 | s | |
| C 15 | 23.2 | 23.87 | CH3 | 0.96 | s | C 14, C 11, C 10, C 1 |
A Molecular Connectivity Diagram (MCD) was created automatically by the program and shown in Figure 1.
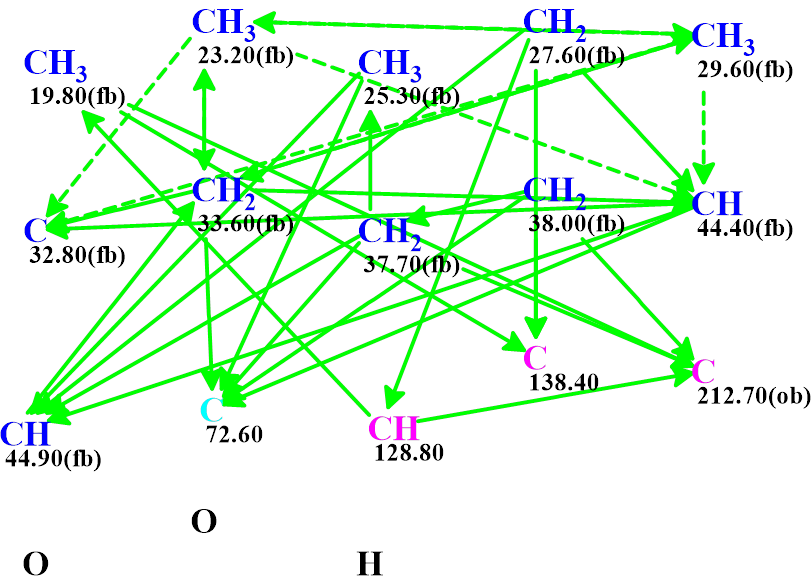
Figure 1. Molecular connectivity diagram. HMBC correlations are marked by green arrows. The hybridizations of carbon atoms are marked by the corresponding colors: sp2 – violet, sp3 – blue, sp2 or sp3 – light blue. Labels “ob” and “fb” are set by the program to carbon atoms for which neighboring with heteroatom is either obligatory (ob) or forbidden (fb).
Logical analysis of the 2D NMR data presented in the MCD showed that no contradictions were detected, therefore strict structure generation accompanied with 13C chemical shift prediction and structural filtering was initiated. As a result, a single structure identical to that suggested by the authors [1] was generated in 0.5 s. Figure 2 shows the structure along with the results of 13C chemical shift prediction using the empirical methods implemented into ACD/SE.
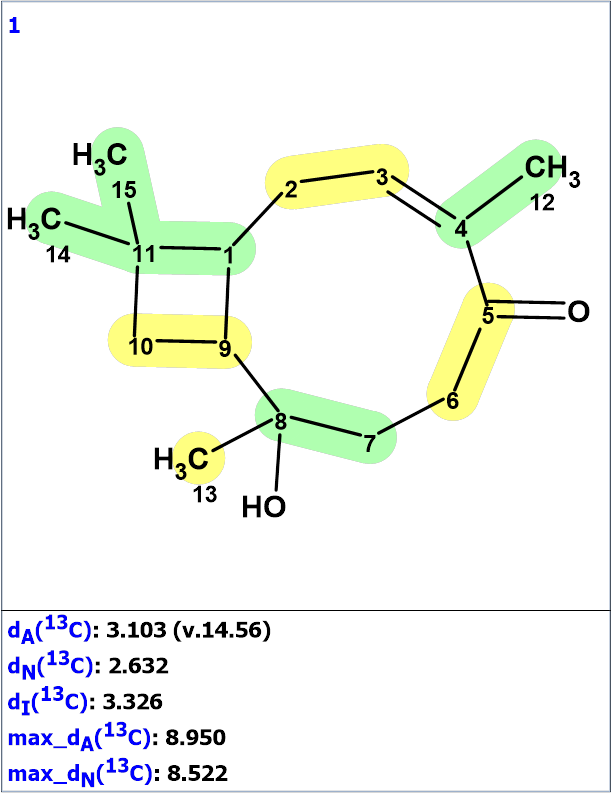
Figure 2. The generated structure. 13C chemical shift prediction was carried out using the HOSE code-based method, neural networks, and the incremental approach. The average deviations of 13C chemical shifts determined by these methods are denoted as dA, dN and dI correspondingly. Each atom is colored to mark a difference between its experimental and calculated 13C chemical shifts. The green color represents a difference between 0 to 3 ppm while yellow indicates a difference of >3 up to 15 ppm.
Although the average deviation values are somewhat higher than those that usually occur for the correct structures determined by the program, the fact that a single structure was derived from the rich HMBC spectrum indicates its correctness.
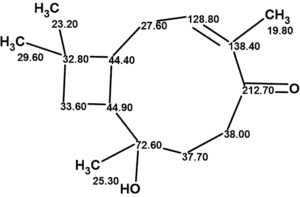
The structure of rumphellol A with assigned 13C chemical shifts is shown below:
References
- H.-M. Chung , W-H. Wang , T.-L. Hwang , J.-J. Chen, L.-S. Fang, Z.-H. Wen, Y.-B. Wang, Y.-C. Wu, P.-J. Sung. (2014). Rumphellols A and B, New Caryophyllene Sesquiterpenoids from a Formosan Gorgonian Coral, Rumphella antipathies. Int. J. Mol. Sci., 15, 15679-15688.


