September 1, 2013
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Jatrophalactam
Jatropha curcas Linn. (Euphorbiaceae) is a native of tropical America, but now thrives in many parts of the tropics and subtropics in Africa/Asia. It is a plant with many attributes, multiple uses, and considerable potential. The oil from J.curcas seeds can be used externally for the treatment of sciatica, dropsy, paralysis, rheumatism, and certain skin diseases. The seeds contain 30% oil that can be processed to produce a high-quality biodiesel fuel, usable in a standard diesel engine. Previous investigations on the genus Jatropha revealed that diterpenoid was their major secondary metabolite, and some of the diterpenoids were cytotoxic and tumor-inhibitory constituents. Wang and co-workers [1] isolated Jatrophalactam (1), a novel diterpenoid lactam possessing an unprecedented skeleton from the roots of Jatropha curcas.
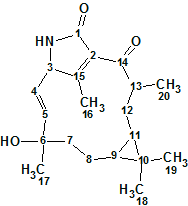
1
Compound 1 was obtained as a colorless crystal. Its molecular formula, C20H29NO3, was established on the basis of HREIMS for the [M]+ at m/z 331.2153, which indicated seven degrees of unsaturation. The IR spectrum showed characteristic absorptions for OH /NH (3392 cm-1, 3266 cm-1) and C=O (1705 cm-1) functions. The 13C NMR of 1 showed 20 carbon signals that were sorted by DEPT experiments into two carbonyls, four olefinic carbons, five methyls, three methylenes, four methines, and two quaternary carbons.
In the HSQC spectrum, a proton at δH 7.00 (brs), which had no correlations with the C-atom, was assigned by authors’ [1] to be an active hydrogen of the NH group.
1D and HSQC data, as well as key COSY and HMBC correlations which were displayed graphically in article [1] are presented in Table 1.
Table 1. Jatrophalactam. Spectroscopic NMR data.
| Label | δC | δCcalc | CHn | δH | M(J) | COSY | C HMBC |
| C 1 | 171.9 | 171.18 | C | ||||
| C 2 | 131 | 132.8 | C | ||||
| C 3 | 60.3 | 62.62 | CH | 4.56 | u | 5.75 | C 1, C 5, C 15, C 4 |
| C 4 | 123.8 | 124.74 | CH | 5.75 | u | 5.84, 4.56 | |
| C 5 | 138.1 | 142.02 | CH | 5.84 | u | 5.75 | C 3 |
| C 6 | 73.7 | 71.03 | C | ||||
| C 7 | 42.2 | 40.78 | CH2 | 1.77 | u | ||
| C 7 | 42.2 | 40.78 | CH2 | 1.33 | u | 1.58 | |
| C 8 | 20 | 16.68 | CH2 | 1.58 | u | 1.33, 0.53 | |
| C 9 | 27 | 20.91 | CH | 0.53 | u | 0.43, 1.58 | |
| C 10 | 18.4 | 18.79 | C | ||||
| C 11 | 21.8 | 20.76 | CH | 0.43 | u | 1.42, 0.53 | |
| C 12 | 28.4 | 27.95 | CH2 | 1.42 | u | 0.43, 2.45 | |
| C 12 | 28.4 | 27.95 | CH2 | 1.48 | u | ||
| C 13 | 47.7 | 45.3 | CH | 2.45 | u | 1.42 | C 14 |
| C 14 | 205.8 | 201.39 | C | ||||
| C 15 | 157.4 | 163.44 | C | ||||
| C 16 | 14.7 | 14.16 | CH3 | 2.01 | u | C 1, C 14, C 3, C 2 | |
| C 17 | 21.1 | 28.82 | CH3 | 1.28 | u | C 6, C 5, C 7 | |
| C 18 | 29.4 | 23.04 | CH3 | 0.98 | u | C 9, C 19, C 10, C 11 | |
| C 19 | 15.6 | 9.64 | CH3 | 0.89 | u | C 11, C 18, C 10 | |
| C 20 | 13.7 | 17.07 | CH3 | 1.26 | u | C 13, C 14, C 12 | |
| N 1 | 100* | NH | 7.00 | u |
* Fictitious 15N chemical shift.
The molecular Connectivity Diagram (MCD), which visualizes the 1D and 2D NMR data in ACD/Structure Elucidator (as collected in Table 1) is presented in Figure 1.

Figure 1. Jatrophalactam Molecular Connectivity Diagram.
No edits of the initial MCD were made. Checking the MCD for contradictions revealed the presence of at least one nonstandard connectivity in the COSY and HMBC data. The first run of the program was performed in the Fuzzy Structure Generation mode, with Determine Option Automatically selected. The results were: k=74→74→41, tg = 0.8s, with 1 from 27 connectivities having been extended during generation, and 4 from 27 possible connectivity combinations used. The two top structures of the ranked output file are presented in Figure 2.

Figure 2. Jatrophalactam. Two top structures of the ranked output file. A red arrow indicates a nonstandard connectivity.
Comparison of structure #1 with the ranked output structure shows that the best solution is correct. A red arrow corresponds to the HMBC connectivity which was automatically elongated by one chemical bond during the Fuzzy Structure Generation. However the average and maximum deviations are relatively large, which, in general, could be explained by an unprecedented skeleton of the molecule. Other causes can be in incorrect 13C chemical shift assignment, accounted for by the possible presence of more than one nonstandard connectivity, as well as by the NSC length of 5 or 6 bonds (parameter a=2 or 3). It is also conceivable, from the common characteristic chemical shifts, that carbon atoms 157.4 and 171.9 should be exchanged in structure #1. To check this hypothesis, the Fuzzy Structure Generation was run again with the following parameters: m = 2, a=1. Results: k = 330→330→142, tg =0.7s, with 2 from 27 connectivities extended during generation, and 98 of 351 possible connectivity combinations were used. The two top structures of the ranked output file are presented in Figure 3.

Figure 3. Jatrophalactam. Two top structures of the ranked output file. Two connectivities have been extended during structure generation (m=2). Red arrows indicate nonstandard HMBC connectivities.
We see that the best structure is still the same, but the average deviations are lesser and indeed carbon atoms 157.4 and 171.9 were automatically exchanged. The same structure with identical chemical shift assignment was obtained when the Fuzzy Structure Generation was repeated with parameters m=2, a=16 (number of nonstandard connectivities is 2, the lengths are unknown). A question arises: whether signals corresponding to nonstandard connectivities in HMBC spectrum could be detected from the very beginning? That question can be answered by inspecting a part of the HMBC pattern (Figure 4) borrowed from Supporting Information to the article [1].

Figure 4. A part of HMBC spectrum for Jatrophalactam. 4JCH correlations from 2.01 to 171.9 and 205.8 are shown in the red circles. Standard length correlations from 4.56 to C1, C4, C5, and C15 of the same intensity are underlined by green lines for comparison.
Figure 4 shows that peak intensifies corresponding to 4JCH correlations (in red circles) are the same as were measured for standard correlations 2-3JCH (examples are underlined by green lines).
Therefore the mechanism of Fuzzy Structure Generation allowed us to instantaneously and automatically reveal the contradictory connectivities, elongate them and find the right structure.
For completeness, we repeated the MCD checking for contradiction with the option Automatically Resolve Contradictions selected. The program distinguished the carbon atom C 14.70 as a potent carrier of connectivity set among which at least one NSC may present. The automatically modified MCD is displayed in Figure 5.

Figure 5.Automatically modified Molecular Connectivity Diagram for Jatrophalactam. Connectivities elongated by one chemical bond were marked by the program with violet color.
Figure 5 shows that both 4JCH correlations from 2.01 to 171.9 and 205.8 were automatically elongated by one chemical bond, and consequently we can expect that the right structure with correct 13C and 1H chemical shift assignments will be generated. As expected, the edited MCD does not contain nonstandard connectivities now, so the Strict Structure Generation was initiated. Results: k=147→147→54, tg = 0.2 s. The top structures and their chemical shift assignments fully coincided with those presented in Figure 3.
The example demonstrates two different methods of resolving the problem of contradictions in 2D NMR data. As has been shown [2] the Fuzzy Structure Generation is more general approach, but there are 2D NMR data sets for which the program manages to find and resolve contradictions at the MCD checking stage.
References
- X.-C. Wang, Z.-P. Zheng, X.-W. Gan, L.-H. Hu., Jatrophalactam, A Novel Diterpenoid Lactam Isolated from Jatropha curcas, Org. Lett., 11(23):5522-5524, 2009.
- M.E. Elyashberg, A.J. Williams, Blinov K.A. Contemporary Computer-Assisted Approaches to Molecular Structure Elucidation, Cambridge, RSC Publishing, 2012. http://pubs.rsc.org/en/Content/eBook/978-1-84973-432-5


