March 1, 2015
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Laevinoid A
The genus Croton (Euphorbiaceae) consists of nearly 1300 species widely distributed in the tropical and subtropical regions all over the world. Many Croton species have long been used as natural remedies in Africa, Asia, and South America, for the treatment of a number of ailments such as cancer and hypertension.
Wang and coworkers [1] collected the branches and leaves of Croton laevigatus from Hainan Island and performed an intensive chemical analysis. They isolated and elucidated structures of two diterpenoids, laevinoids A (1) and B, which represent a new rearranged ent-clerodane scaffold with an unusual 3/5 bicyclic motif. The absolute structures of the compounds were elucidated via MS, NMR, and X-ray crystallographic data.
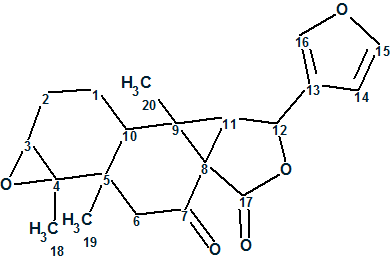
1
Laevinoid A (1) possessed a molecular formula of C20H22O5 with 10 degrees of unsaturation, as established by HRESI(-)MS analysis at m/z 387.1434 ([M + HCO2]–, calculated 387.1444). The IR spectrum indicated the presence of lactone (1763 cm-1) and ketone (1691 cm-1) functionalities. The 1H NMR data of 1 displayed diagnostic signals for three tertiary methyls (dH 1.17, 1.20, and 1.26) and a typical β-substituted furan ring (dH 6.39, 7.41, and 7.48). The 13C NMR data (CDCl3) revealed 20 carbon resonances that were distinguished via DEPT and HSQC data to be three methyls, three methylenes, seven methines, and seven quaternary carbons. The above-mentioned analysis accounted for 5 out of 10 degrees of unsaturation, indicating five additional rings in the structure of 1 besides the furan ring.
The spectroscopic data for Laevinoid A are presented in Table 1, where a column Ccalc contains chemical shifts predicted by HOSE code-based algorithm.
Table 1: NMR spectroscopic data for Laevinoid A.
| C Label | δC | δC Calc | CHn | δH | M | COSY | C HMBC |
| C 1 | 19.4 | 19.04 | CH2 | 1.58 | u | 1.73, 1.04 | |
| C 1 | 19.4 | 19.04 | CH2 | 1.69 | u | ||
| C 2 | 26.3 | 24.05 | CH2 | 1.73 | u | 3.02, 1.58 | |
| C 2 | 26.3 | 24.05 | CH2 | 2.22 | u | ||
| C 3 | 61.6 | 59.14 | CH | 3.02 | u | 1.73 | |
| C 4 | 61.7 | 63.03 | C | ||||
| C 5 | 39.4 | 42 | C | ||||
| C 6 | 50.9 | 46.66 | CH2 | 2.08 | u | ||
| C 6 | 50.9 | 46.66 | CH2 | 2.55 | u | C 7, C 8 | |
| C 7 | 200.4 | 205.47 | C | ||||
| C 8 | 43.5 | 44.6 | C | ||||
| C 9 | 34.6 | 33.09 | C | ||||
| C 10 | 50.3 | 41.99 | CH | 1.04 | u | 1.58 | |
| C 11 | 49.4 | 39.82 | CH | 2.5 | u | 5.2 | C 17, C 7 |
| C 12 | 70.5 | 74.13 | CH | 5.2 | u | 2.5 | C 17, C 8, C 13, C 16 |
| C 13 | 123.6 | 121.22 | C | ||||
| C 14 | 108 | 107.9 | CH | 6.39 | u | 7.41 | |
| C 15 | 144.5 | 143.66 | CH | 7.41 | u | 6.39 | |
| C 16 | 140.2 | 139.45 | CH | 7.48 | u | ||
| C 17 | 169.6 | 170.36 | C | ||||
| C 18 | 19.9 | 16.95 | CH3 | 1.17 | u | C 5, C 3, C 4 | |
| C 19 | 15.8 | 19.95 | CH3 | 1.2 | u | C 10, C 6, C 4, C 5 | |
| C 20 | 10.8 | 19.58 | CH3 | 1.26 | u | C 8, C 9, C 11, C 10 |
This data was input into ACD/Structure Elucidator Suite, which produced the following Molecular Connectivity Diagram (MCD):
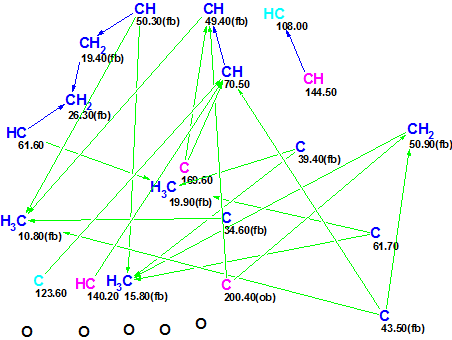
Figure 1: Initial Molecular Connectivity Diagram.
The diagram shows that two carbons, CH 108.0 and C 123.6, are marked in a light blue color. This means that the hybridization of these two atoms is set by the program as “not sp” (sp2 or sp3). It is probable that the sp3 carbons 61.7, 61.6 and 70.5 are oxygenated, and C 169.6 belongs to a carbonyl, but no edits of the initial MCD were made.
Structure generation combined with the 13C chemical shift prediction was performed from the MCD. The structures for which average deviations between experimental and calculated values exceed 5 ppm were eliminated during the structure generation process. The following results were obtained: k=5979→1, tg = 17s. The single structure delivered by the program (Figure 2) was identical to that suggested by authors [1].
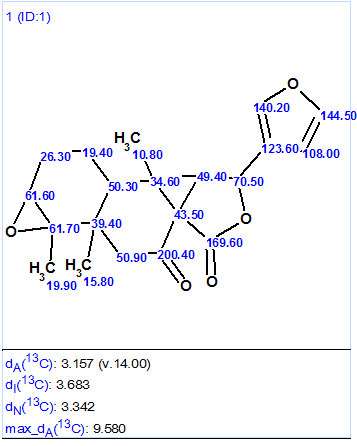
Figure 2. The single structure generated by the software’s first run.
The values of all average deviations are relatively large, which is explained by the unique features of the molecule’s unusual skeleton.
The original authors [1] also performed an X-ray diffraction experiment, which allowed them the to determine the stereochemistry of the molecule as shown in structure 2:
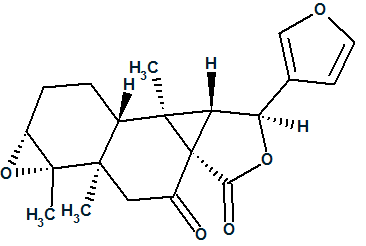
2
In earlier work [2] it has been shown that 13C chemical shift predictions using HOSE-code based algorithms can be used as a preliminary filter for selecting the most probable Laevinoid A stereoisomers from among the full set of possibilities. With this in mind, 256 stereoisomers were generated and ranked in increasing order of deviation. The six top-ranked stereoisomers are presented in Figure 3.
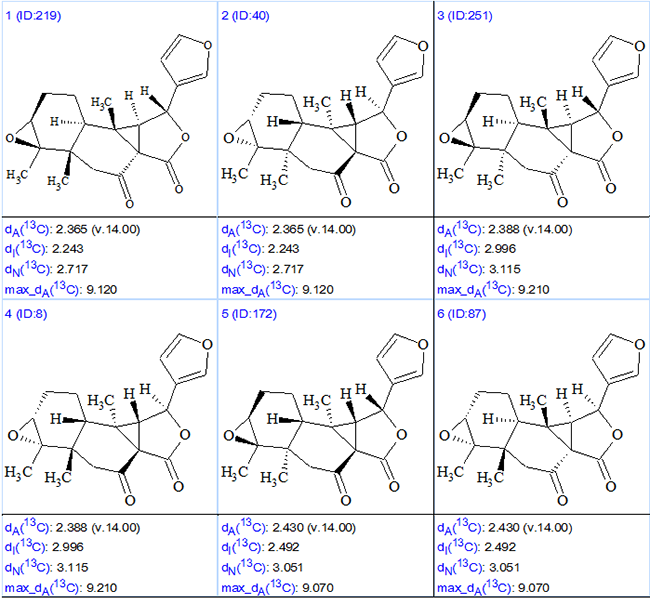
Figure 3. The six top ranked stereoisomers for Laevinoid A.
Two first ranked structures are antipodes. It is evident that stereoisomer #2 has a configuration similar to that determined by the original authors [1]. To determine if these stereoisomers are identical, for both stereoisomeric structures (2 and #2) stereocenters were classified using a corresponding ChemSketch option (Tools/Generate/InChI for Structure) and InChI codes were generated as shown in Figure 4.
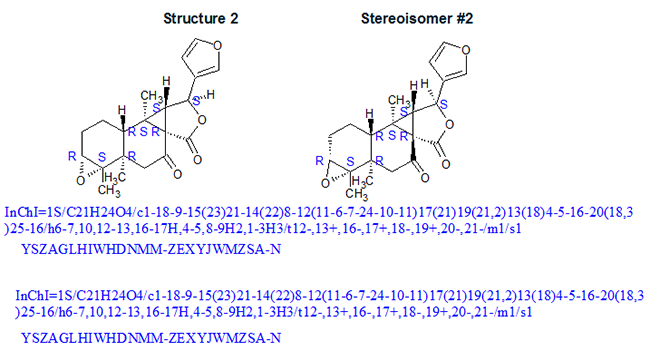
Figure 4. Comparison of stereoisomers (Structure 2 and Stereoisomer #2).
Figure 4 demonstrates that InChI codes of both stereo configurations are identical.
Therefore Structure Elucidator Suite not only determined structure of an unknown compound characterized by unique skeleton, but also correctly selected the most probable stereoisomer, without the need for editing the MCD or any further adjustments in the structure generation process.
References
- G-C. Wang, H. Zhang, H.-B. Liu and J.-M. Yue. (2013). Laevinoids A and B: Two Diterpenoids with an Unprecedented Backbone from Croton laevigatus, Org. Lett. 15:4880-4883.
- Elyashberg, M.E.; Blinov, K.; Williams, A.J. (2009). The Application of Empirical Methods of NMR Chemical Shift Prediction as a Filter for Determining Possible Relative Stereochemistry, Magn. Reson. Chem., 47:333-341


