December 1, 2017
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Manginoid A
The enzyme 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) metabolizes inactive glucocorticoids (GCs). These are known to act as functional antagonists of insulin; therefore, it is desirable to have molecules that could act as inhibitors of 11β-HSD1. There have been many natural products found inhibiting against 11β-HSD1 but most have had rather low such activity. It is easy to understand why there is an interest in developing better inhibitors.
Meroterpenoids have always been interesting for chemists due to the high variety of their structures and their biological activity. A series of meroterpenoids has already been isolated from G. mangiferae, named guignardones. In the study conducted by Chen et al [1] manginoids A−G were isolated from G. mangiferae. These were seven monoterpene-shikimate conjugated meroterpenoids related to guignardones. Their structures were elucidated as part of that study. Particularly manginoids A-D are the first meroterpenoids uniquely defined by the presence of a 6-oxaspiro[bicyclo[3.2.1]octane-3,5′-indene] skeleton. Of all those isolated, manginoid A exhibits outstanding inhibitory effect against 11β-HSD1.
Manginoid A (1) was assigned the molecular formula C17H22O5 based on its HR-ESI-MS spectrum, with a sodium adduct molecular ion peak at m/z 329.1378 ([M + Na]+, calculated for C17H22O5Na, 329.1365).

1
Spectroscopic data used for the structure elucidation of Manginoid A were used to challenge ACD/Structure Elucidator Suite. The NMR spectroscopic data including only key HMBC and COSY correlations available from the article [1] are collected in Table 1.
Table 1. Manginoid A. 1D and 2D NMR spectroscopic data
| Label | δC | δC calc (HOSE) |
CHn | δH | COSY | H to C HMBC |
|---|---|---|---|---|---|---|
| C 1 | 81.1 | 81.68 | C | |||
| C 2 | 209.2 | 208.76 | C | |||
| C 3 | 61.1 | 64.78 | C | |||
| C 4 | 205.2 | 205.09 | C | |||
| C 5 | 81.7 | 82.58 | CH | 4.61 | 2 | C 1, C 3, C 4 |
| C 7 | 71.3 | 72.32 | CH2 | 3.93 | C 1, C 5, C 8 |
|
| C 7 | 71.3 | 72.32 | CH2 | 3.91 | ||
| C 8 | 35.7 | 48.93 | CH2 | 2 | 4.61 | C 1, C 2, C 4 |
| C 8 | 35.7 | 48.93 | CH2 | 2.72 | ||
| C 9 | 29.5 | 28.74 | CH2 | 2.16 | 2.01 | C 2, C 3, C 4 |
| C 9 | 29.5 | 28.74 | CH2 | 1.61 | ||
| C 10 | 51.4 | 53.91 | CH | 2.01 | 2.16, 2.30 |
|
| C 11 | 78.9 | 80.62 | C | |||
| C 12 | 40 | 39.98 | CH2 | 1.81 | ||
| C 12 | 40 | 39.98 | CH2 | 1.9 | 1.85 | |
| C 13 | 23.9 | 26.61 | CH2 | 1.64 | ||
| C 13 | 23.9 | 26.61 | CH2 | 1.85 | 1.90, 2.30 |
|
| C 14 | 46.1 | 49.67 | CH | 2.3 | 1.85, 2.01 |
|
| C 15 | 146 | 147.4 | C | |||
| C 16 | 44.6 | 39.69 | CH2 | 2.43 | ||
| C 16 | 44.6 | 39.69 | CH2 | 2.56 | C 2, C 3, C 4, C 9 |
|
| C 17 | 26.7 | 23.82 | CH3 | 1.27 | C 10, C 11, C 12 |
|
| C 18 | 107.4 | 111.72 | CH2 | 4.75 | C 14, C 15, C 16 |
|
| C 18 | 107.4 | 111.72 | CH2 | 4.64 |
The Molecular Connectivity Diagram (MCD) automatically created by the program from Table 1 is shown in Figure 1.
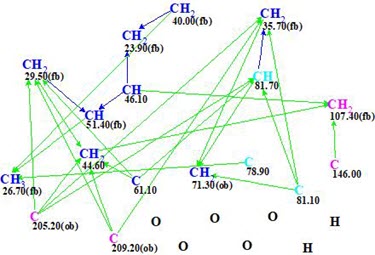
Figure 1. Molecular connectivity diagram.
No manual MCD edits were performed, and structure generation was initiated. As a result only one structure was produced in 0.06 s (Figure 2).
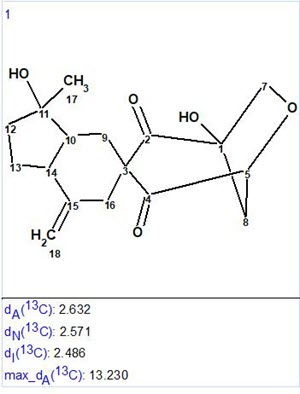
Figure 2. Resulting structure with average 13C chemical shift deviations.
The structure obtained coincides with the structure of Manginoid A determined by the authors of [1]. Values of average deviations are common for a correct structure, although the maximum deviation found for HOSE code based 13C chemical shift calculation is relatively large (13.2 ppm). This value comes from the CH2 (C-8) situated on the top of a bridge. To explain this the protocol of the HOSE code chemical shift calculation was analyzed. It was established that only two structures similar enough and appropriate for 13C chemical shift prediction for C-8 were present in the ACD/CNMR database (Figure 3).
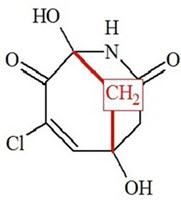
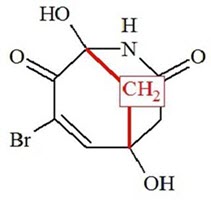
Figure 3. Two structures existing in the ACD/CNMR database used for C-8 chemical shift prediction. 13C chemical shifts of CH2 groups (red) are around 47 ppm.
We see that only the one bond length environment corresponds to structure 1, and this is the reason for the inaccurate chemical shift prediction (see Table 1).
A fast and unique solution to the problem was found because the molecule under investigation is small enough and 2D NMR data are complete [1]. It was interesting to see what would happen if the COSY data are excluded from analysis. In this case, structure generation gave the following result: k=228→107→106, tg =0.3 s. The top structures of the output file are shown in Figure 4.
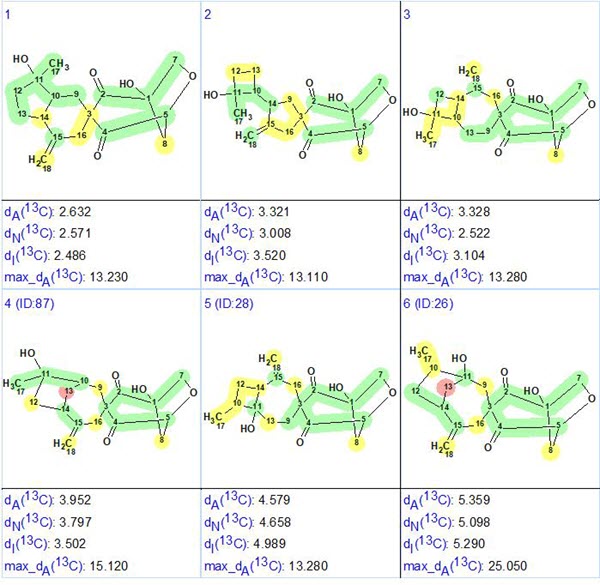
Figure 4. Six top ranked structures of the output file obtained when COSY data were switched off.
We see that the correct structure was again selected confidently by the ranking procedure.
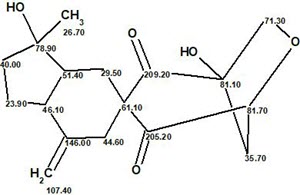
Finally the structure of Manginoid A with the 13C chemical shift assignment carried out by the program is shown below:
A complete elucidation package that speeds up the elucidation process and ensures that no candidate is overlooked. Learn more about ACD/Structure Elucidator Suite.
References:
- K. Chen, X. Zhang, W. Sun, J. Liu, J. Yang, C. Chen, X. Liu, L. Gao, J. Wang, H. Li, Z. Luo, Y. Xue, H. Zhu, Y. Zhang. (2017). Manginoids A−G: Seven Monoterpene−Shikimate-Conjugated Meroterpenoids with a Spiro Ring System from Guignardia mangiferae. Org. Lett., 19, 5956−5959, DOI: 10.1021/acs.orglett.7b02955
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.


