May 1, 2016
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Mannolide A
Cephalotaxus is the only genus of the Cephalotaxaceae family, and is well-known for its interesting secondary metabolites such as alkaloids and terpenoids with a variety of biological properties, particularly antitumor activity. Among the Cephalotaxus metabolites, the troponoids represent a rare class of C19 norditerpenoids incorporating a highly rigid tetracyclic carbon skeleton. Ni et al [1] investigated for the first time Cephalotaxus mannii Hook f. that grows in Xishuangbanna of China, which led to the isolation of three new diterpenoids, namely, mannolides A−C, and two new Cephalotaxus troponoids. Mannolides A−C feature a new intact C20 diterpenoid skeleton for which the authors [1] proposed a trivial name of cephalotane.
We used the example of Mannolide A (1) to challenge our ACD/Structure Elucidator Suite software.
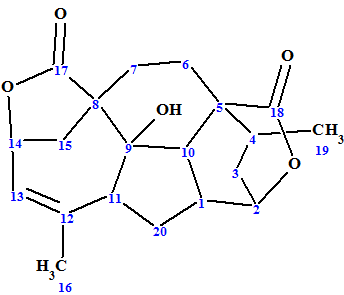
1
Mannolide A was obtained as a white solid. Its molecular formula was determined to be C20H24O5 by HRESIMS at m/z 711.3162 [2M + Na]+ (calculated for C40H48O10Na, 711.3145), and implied nine double bond equivalents. The IR spectrum of 1 revealed the presence of a hydroxyl group (3485 cm−1) and two lactone carbonyls (1755 and 1738 cm−1). The 1D and 2D NMR data used by authors [1] for Mannolide A structure determination are presented in Table 1.
Table 1. Spectroscopic 1D and 2D NMR data for Mannolide A.
| Label | δC | δC* calc | CHn | δH | COSY | C HMBC |
| C 1 | 36.100 | 39.040 | CH | 2.960 | 2.20, 2.33, 4.66 | |
| C 2 | 77.800 | 76.940 | CH | 4.660 | 2.18, 2.96 | C 18 |
| C 3 | 30.000 | 32.430 | CH2 | 2.180 | 2.08, 4.66 | |
| C 3 | 30.000 | 32.430 | CH2 | 1.610 | ||
| C 4 | 31.700 | 32.690 | CH | 2.080 | 0.97, 2.18 | |
| C 5 | 43.700 | 47.810 | C | |||
| C 6 | 22.000 | 27.620 | CH2 | 2.510 | 2.40 | C 4, C 5, C 18 |
| C 6 | 22.000 | 27.620 | CH2 | 1.510 | ||
| C 7 | 27.900 | 26.810 | CH2 | 2.400 | 2.51 | C 5, C 9, C 17 |
| C 7 | 27.900 | 26.810 | CH2 | 1.510 | ||
| C 8 | 50.300 | 52.480 | C | |||
| C 9 | 82.900 | 80.770 | C | |||
| C 10 | 50.800 | 49.570 | CH | 2.200 | 2.96 | C 5, C 6, C 8, C 9, C 11, C 18 |
| C 11 | 49.100 | 46.610 | CH | 3.200 | 2.33 | |
| C 12 | 142.400 | 138.040 | C | |||
| C 13 | 121.800 | 122.990 | CH | 5.760 | 4.98 | C 15 |
| C 14 | 75.500 | 74.410 | CH | 4.980 | 2.71, 5.76 | C 12, C 17 |
| C 15 | 38.700 | 40.230 | CH2 | 2.260 | ||
| C 15 | 38.700 | 40.230 | CH2 | 2.710 | 4.98 | C 8, C 9, C 17 |
| C 16 | 24.100 | 23.190 | CH3 | 1.850 | C 11, C 12, C 13 | |
| C 17 | 178.600 | 181.650 | C | |||
| C 18 | 175.500 | 175.780 | C | |||
| C 19 | 20.800 | 16.960 | CH3 | 0.970 | 2.08 | C 5 |
| C 20 | 28.100 | 33.020 | CH2 | 2.330 | 2.96, 3.20 | C 2, C 9, C 12 |
| C 20 | 28.100 | 33.020 | CH2 | 1.710 |
* 13C chemical shift calculations were carried out using a HOSE code based approach [2]
The data collected in Table 1 were input into ACD/Structure Elucidator Suite, and a Molecular Connectivity Diagram (MCD) [2] was created (Figure 1).
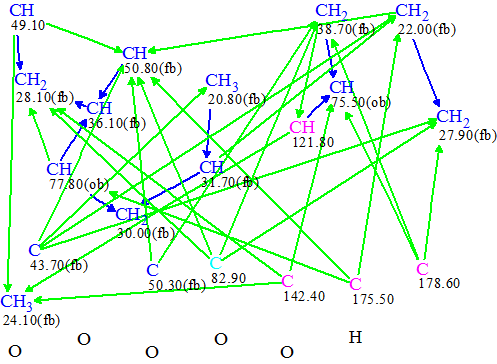
Figure 1. Molecular Connectivity Diagram for Mannolide A.
Figure 1 shows that carbon atoms are involved in a dense net of HMBC and COSY connectivities. All sp3-hybridized carbon atoms (blue) were marked by the program as those which either are connected to a neighboring heteroatom (“ob” or obligatory) or not connected (“fb” or forbidden). Therefore, in spite of the fact that all sp2-hybridized carbon atoms (violet) received no such labels, and the hybridization of a blue carbon (C82.9) was set ambiguously as sp3 or sp2, we expected that structure generation would not take a lot of time. Thus no MCD edits were made, and Strict Structure Generation was initiated, resulting in a single unique structure being generated in 0.07s. The calculated average
deviations for the unique structure are shown below:
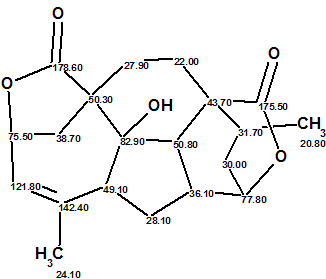
This unique structure coincides with the structure of Mannolide A suggested by the original authors [1]; the predicted chemical shift deviations are dA=2.26, dN=2.62, dI =2.42 ppm.
Thus, the correct structure of Mannolide A (confirmed by x-ray crystallography [1]) was unequivocally and fully automatically determined in no time using ACD/Structure Elucidator Suite. The quick success was accounted for by the fact that the software could produce a comprehensive set of initial axioms from the experimental data which (a large number of HMBC and COSY correlations were observed) and found no contradictions were present in the 2D NMR data.
References
- G. Ni, H. Zhang, Y.-Y. Fan, H.-C. Liu, J. Ding, J.-M. Yue. Mannolides A−C with an Intact Diterpenoid Skeleton Providing Insights on the Biosynthesis of Antitumor Cephalotaxus Troponoids. Org. Lett., ASAP, Received: March 7, 2016, DOI: 10.1021/acs.orglett.6b00653.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.


