October 1, 2017
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Matsutakone—Pleurocin A
A well-known source of natural products are mushrooms. They are rich in organic compounds with sophisticated skeletons and/or promising bioactivities. The mycorrhizal fungus Tricholoma matsutake is a kind of very delicious and nutritious wild mushroom from China. As such it is often used in Chinese cooking. It can be found in the roots of pine and oak trees. Previous studies showed that the major contents of T. matsutake were crude fat, protein, and polysaccharides. However, few of the secondary metabolites from T. matsutake have been reported. Zhao et al [1] reported a chemical investigation on natural products of T. mastutake aimed at discovering potential drug leads that led to the isolation of two novel and one known steroid.
The novel matsutakone 1 represented a new class of steroids with an unprecedented polycyclic system.
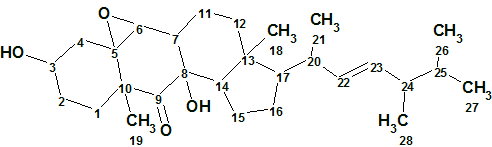
1
The ethyl acetate layer of T. matsutake (2.0 kg) was fractionated and purified by repeated chromatographic methods to yield 1 (1.4 mg). The white powder matsutakone (1) was assigned the chemical formula C28H44O4 (seven degrees of unsaturation) by a positive-ion HRMS (ESI-TOF) experiment m/z: [M + Na]+ calcδ for C28H44O4Na 467.3132; found 467.3121.
Structure of 1 was elucidated from its molecular formula and 1D and 2D NMR data shown in Table 1, which were used by us for challenging ACD/Structure Elucidator.
Table 1. Spectroscopic 1D and 2D NMR data
| C/X Label | δC | δC calc | XHn | δH | COSY | H to CHMBC |
| C 1 | 30.8 | 27.5 | CH2 | 2.15 | 2 | |
| C 1 | 30.8 | 27.5 | CH2 | 2.1 | ||
| C 2 | 31.5 | 30.32 | CH2 | 2.26 | ||
| C 2 | 31.5 | 30.32 | CH2 | 2 | 2.15, 4.47 |
|
| C 3 | 68.3 | 68.35 | CH | 4.47 | 1.84, 2.00 |
|
| C 4 | 39.5 | 37.3 | CH2 | 2.48 | ||
| C 4 | 39.5 | 37.3 | CH2 | 1.84 | 4.47 | |
| C 5 | 69.8 | 63.41 | C | |||
| C 6 | 63.7 | 60.44 | CH | 3.39 | 2.58 | C 4 |
| C 7 | 49.5 | 46.2 | CH | 2.58 | 1.83, 3.39 |
|
| C 8 | 75.9 | 76.95 | C | |||
| C 9 | 213 | 211.41 | C | |||
| C 10 | 47.2 | 50.99 | C | |||
| C 11 | 25.7 | 22.07 | CH2 | 2.1 | ||
| C 11 | 25.7 | 22.07 | CH2 | 1.83 | 1.94, 2.58 |
C 8 |
| C 12 | 40.1 | 34.08 | CH2 | 1.94 | 1.83 | C 7 |
| C 12 | 40.1 | 34.08 | CH2 | 1.28 | ||
| C 13 | 43.9 | 45.87 | C | |||
| C 14 | 60.3 | 54.11 | CH | 1.87 | H 14 | |
| C 15 | 20.9 | 23.77 | CH2 | 2.66 | 1.46 | C 8, C 14 |
| C 15 | 20.9 | 23.77 | CH2 | 1.99 | ||
| C 16 | 29.1 | 28.31 | CH2 | 1.46 | 1.20, 2.66 |
|
| C 16 | 29.1 | 28.31 | CH2 | 1.74 | ||
| C 17 | 52.7 | 56.6 | CH | 1.2 | 1.46, 2.06 |
|
| C 18 | 14.9 | 20.63 | CH3 | 0.99 | C 12, C 13, C 14, C 17 |
|
| C 19 | 23.4 | 18.68 | CH3 | 1.77 | C 1, C 5, C 9, C 10 |
|
| C 20 | 40.8 | 39.39 | CH | 2.06 | 1.05, 1.20, 5.20 |
|
| C 21 | 21.5 | 20.76 | CH3 | 1.05 | 2.06 | |
| C 22 | 136.5 | 136.15 | CH | 5.2 | 2.06, 5.26 |
|
| C 23 | 132.4 | 132.27 | CH | 5.26 | 1.87, 5.20 |
|
| C 24 | 43.4 | 43.14 | CH | 1.87 | ||
| C 25 | 33.7 | 33.52 | CH | 1.47 | 1.87, 0.85, 0.86 |
|
| C 26 | 20.2 | 19.67 | CH3 | 0.85 | 1.47 | |
| C 27 | 20.5 | 20.1 | CH3 | 0.86 | 1.47 | |
| C 28 | 18.2 | 17.84 | CH3 | 0.94 | 1.87 | |
| O 1 | OH | 6.44 | ||||
| O 2 | OH | 1.87 | 0.94, 1.47, 5.26 |
C 7, C 8, C 9, C 14 |
Figure 1 shows the Molecular Connectivity Diagram (MCD) created by the program from the data in Table 1.
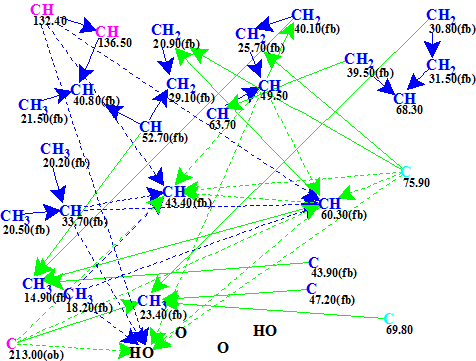
Figure 1. Molecular connectivity diagram (MCD).
MCD overview. Hybridizations of almost all carbon atoms were automatically set by the program. In the MCD carbons with sp3 hybridization are shown in blue color, while sp2 in violet. Two atoms, C 75.9 and C 69.8, are colored in light blue color, which means that their hybridization may be sp3 or sp2. Labels “fb” and “ob” are used to distinguish atoms for which connections to a heteroatom (oxygen in this case) are either forbidden or obligatory, correspondingly. HMBC correlations are shown as green arrows, COSY correlations as blue ones. Dotted lines denote ambiguous connectivities which arise due to some 1H signal overlapping. No manual edits of the MCD were made.
As no contradictions were found in 2D NMR data, a Strict Structure Generation was initiated which gave the following results: k = 726 → 43 → 24, tg = 3 s.
Then, as it is common for CASE methodology [2], 13C chemical shifts were predicted for the structures generated by three empirical methods (incremental, neural networks, and HOSE code based) implemented into ACD/Structure Elucidator. In addition, 1H chemical shifts were predicted using the neural networks method. Finally the structures in the output file were ranked in increasing order of the dA value (average chemical shift deviation calculated by HOSE code based approach). The six top ranked structures of the output file are shown in Figure 2.
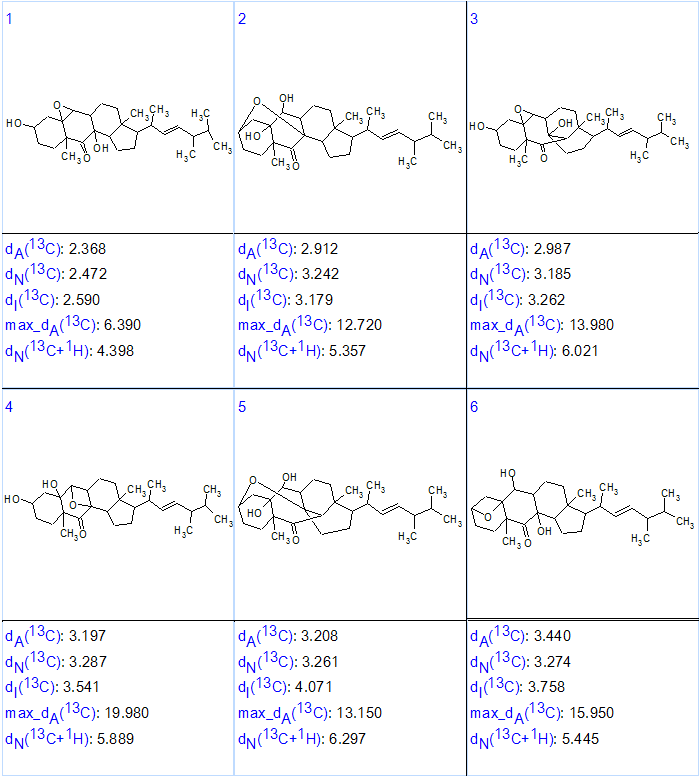
Figure 2. The six top ranked structures of the output file.
We see that all average and maximum deviations indicate that the structure which is identical to matsutakone 1 is really the most probable one with a reasonable margin from the next one. The elucidated structure along with the automatically assigned 13C chemical shifts is shown below.
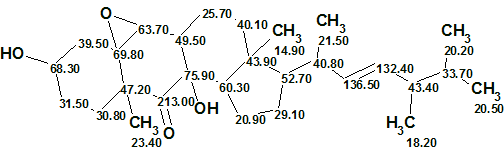
While preparing this article for the Elucidation of the Month it has come to our attention that the group of Reiko Tanaka from the Osaka University of Pharmaceutical Sciences has isolated the exact same compound from the fruiting bodies of Pleurotus eryngii (Pleurotaceae) [5]. They named the compound Pleurocin A. The two reports appeared almost simultaneously (received by the journal on 18th and 23rd May 2017, published on the web on Jul 10th and August 31st respectively), in a very rare case of isolation of the same unknown natural product from two different sources at practically the same time by two different groups.
References
- Z.-Z. Zhao, H.-P. Chen, B. Wu, L. Zhang, Z.-H. Li, T. Feng, J.-K. Liu. (2017) Matsutakone and Matsutoic Acid, Two (Nor)steroids with Unusual Skeletons from the Edible Mushroom Tricholoma matsutake. J. Org. Chem., 82: 7974−7979.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.
- M.E. Elyashberg, K.A. Blinov, A.J. Williams. (2009) The Application of Empirical Methods of 13C NMR Chemical Shift Prediction as a Filter for Determining Possible Relative Stereochemistry. Magn. Reson. Chem., 47: 333-341.
- A. V. Buevich, M. E. Elyashberg. (2016) Synergistic combination of CASE algorithms and DFT chemical shift predictions: a powerful approach for structure elucidation, verification and revision. J. Nat. Prod., 79 (12): 3105–3116.
- T. Kikuchi, Y. Horii, Y. Maekawa, Y. Masumoto, Y. In, K. Tomoo, H. Sato, A. Yamano, T. Yamada, R. Tanaka, Pleurocins A and B: Unusual 11(9 → 7)-abeo-Ergostanes and Eringiacetal B: A 13,14-seco-13,14-Epoxyergostane from Fruiting Bodies of Pleurotus eryngii and Their Inhibitory Effects on Nitric Oxide Production. J. Org. Chem., Article ASAP, DOI: 10.1021/acs.joc.7b01259


