February 1, 2019
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Neosetophomone A
Meroterpenoids are compounds originating from hybrid terpenoid and polyketide biosynthesis. They are interesting because of their unique ring systems and their wide-range of biological activities. Their unusual structures are a result of the enzyme reactions that produce them. A recently described ascomycete genus in the fungi kingdom is Neosetophoma (Phaeosphaeriaceae). A very limited set of secondary metabolites have been reported from Neosetophoma samarorum. El-Elimat et al [1] reported five new and one known meroterpenoids, including the discovery of a novel ioxa[4.3.3]propellane metabolite neosetophomone A (1) that incorporates a 3-methyl-2,3-dihydrofuran bridge into a 5,6 tricyclic 2-hydroxycyclopent-2-en-1-one/tetrahydropyran ring system.
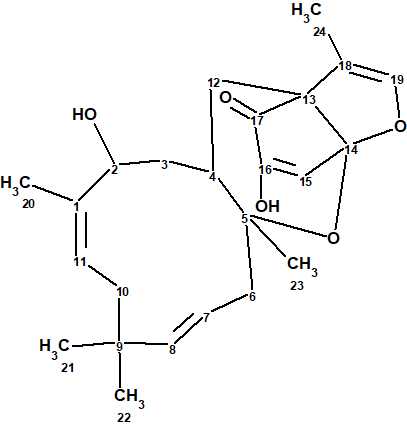
1
Compound 1 (3.94 mg) was isolated as a white amorphous powder. The molecular formula was found by HR-ESI-MS to be C24H32O5, indicating an index of hydrogen deficiency of 9.
NMR spectroscopic data (1D, HSQC, HMBC and COSY) presented in [1] are shown in Table 1.
Table 1. NMR spectroscopic data obtained for compound 1
| Label | δC | δC calca | CHn | δH | COSY | H to C HMBC |
| C 1 | 137.1 | 138.6 | C | |||
| C 2 | 68.5 | 72.22 | CH | 4.59 | 1.42 | C 20, C 4, C 11 |
| C 3 | 38.5 | 36.05 | CH2 | 1.42 | 1.63; 4.59 | C 12, C 4, C 2, C 5, C 1 |
| C 3 | 38.5 | 36.05 | CH2 | 1.94 | ||
| C 4 | 31.5 | 43.1 | CH | 1.63 | 1.26, 1.42 | C 12 |
| C 5 | 81.4 | 79.37 | C | |||
| C 6 | 49.1 | 43.02 | CH2 | 2.38 | 5.26 | C 23, C 4, C 5, C 7, C 8 |
| C 6 | 49.1 | 43.02 | CH2 | 2.49 | C 23, C 4, C 5, C 7, C 8 | |
| C 7 | 121.8 | 120.82 | CH | 5.26 | H 16 | |
| C 8 | 144.2 | 143.44 | CH | 5.26 | 2.38 | C 22, C 9, C 6, C 7 |
| C 9 | 36.3 | 36.98 | C | |||
| C 10 | 42.1 | 38.93 | CH2 | 1.76 | 5.45 | C 22, C 9, C 11, C 1, C 8 |
| C 10 | 42.1 | 38.93 | CH2 | 2.11 | C 22, C 9, C 11, C 1 | |
| C 11 | 125.7 | 123.8 | CH | 5.45 | 1.76 | C 20, C 2 |
| C 12 | 27.8 | 29.19 | CH2 | 2.12 | C 3, C 13, C 5, C 14 | |
| C 12 | 27.8 | 29.19 | CH2 | 1.26 | 1.63 | C 4, C 3, C 13, C 5, C 18, C 17 |
| C 13 | 56.6 | 61.44 | C | |||
| C 14 | 111.1 | 112.24 | C | |||
| C 15 | 123.1 | 123.56 | CH | 6.28 | C 13, C 17 | |
| C 16 | 153.2 | 149.94 | C | |||
| C 17 | 199.3 | 199.81 | C | |||
| C 18 | 109.9 | 114.07 | C | |||
| C 19 | 140.3 | 142.12 | CH | 6.14 | C 24, C 13, C 14 |
|
| C 20 | 19.4 | 18.09 | CH3 | 1.72 | C 2, C 11, C 1 |
|
| C 21 | 28.4 | 27.18 | CH3 | 1.08 | C 22, C 9, C 10, C 8 |
|
| C 22 | 26.8 | 27.18 | CH3 | 1.04 | C 21, C 9, C 10, C 8 |
|
| C 23 | 23.2 | 23.81 | CH3 | 1.17 | C 4, C 6, C 5, C 14 |
|
| C 24 | 8.6 | 11.98 | CH3 | 1.65 | C 13, C 18, C 19, C 17 |
a C13 chemical shifts prediction was performed by HOSE code based empirical method.
The data shown in Table 1 were entered into ACD/Structure Elucidator. The program created automatically the Molecular Connectivity Diagram (MCD) shown in Figure 1.
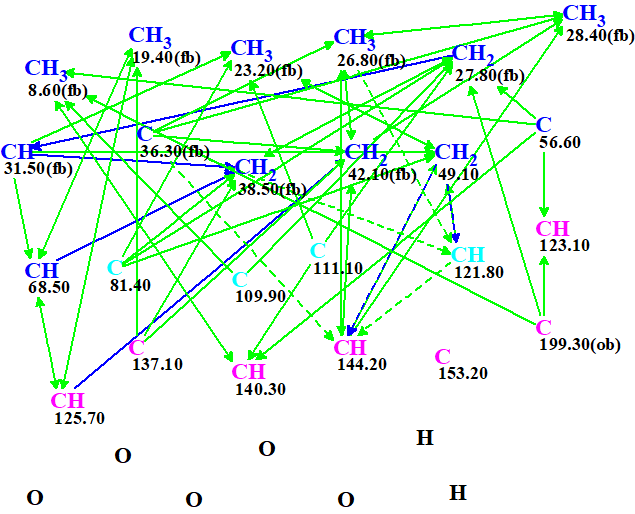
Figure 1. Molecular connectivity diagram.
MCD overview. Four carbon atoms distinguished by light blue color were assigned undefined hybridization “not sp” (sp2 or sp3). Two COSY connectivities are ambiguous due to the presence of two overlapped 1H signals at 5.26 ppm (see Table 1). These connectivities are marked by a blue dotted line. Ten sp3-hybridized atoms were defined as “fb”, which means the presence of a neighboring oxygen atom is forbidden for them. An “ob” label at carbon 199.30 ppm defines a carbonyl group.
No edit of the MCD created by the program was made. The MCD checking for the presence of nonstandard connectivities (NSCs) was completed with the following message:
“The minimum number of non-standard connectivities is 1”.
Therefore Fuzzy Structure Generation (FSG) [2] accompanied with 13C chemical shift prediction by the incremental method and spectral filtering was initiated. Options of FSG were set by the program automatically. Results: k = 1274 → 5 → 5, tg = 5 s; 2 (from 46) connectivities have been extended (deleted) during generation. 1035 (from 1035 possible) connectivity combinations have been used during generation.
We see that the program detected the presence of two NSCs and saved after filtering five structures satisfying this condition in five seconds.
13C chemical shift prediction was carried out for the structures in the output file using two other methods also implemented into ACD/Structure Elucidator – HOSE code and neural networks based. The output file was ranked in increasing order of average deviation dA(13C) values (related to HOSE code based approach). The ranked structural file is shown in Figure 2.
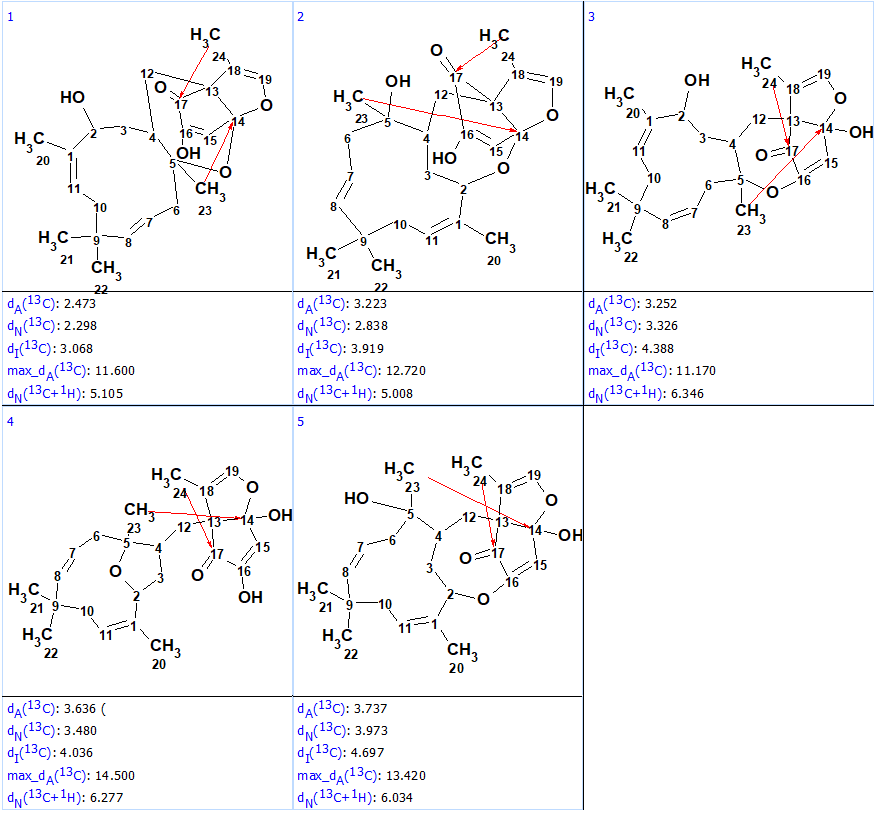
Figure 2. Ranked output file.
Figure 2 shows that the first ranked structure #1 coincides with the structure of neosetophomone A suggested by the authors [1]. It contains two NSCs of three C-C bonds length (4JCH) which are marked with red arrows in structure. As shown in Figure 3 (part of HMBC pattern presented in Supporting Information to [1]) by arrows, the HMBC peaks corresponding to correlations 1.65 / 199.3 and 1.17 / 111.1 are of relatively small intensity, which is in agreement with the lengths of NSCs detected. In the same time, the lengths of NSCs depicted in structures #2 – #5 are extremely large, and this fact additionally confirms the validity of the top structure. Such weak peaks are not unusual at all in HMBC spectra and a strict handling of the dataset would have caused the structure generation to fail. FSG had no problem with it and the structures were successfully generated.
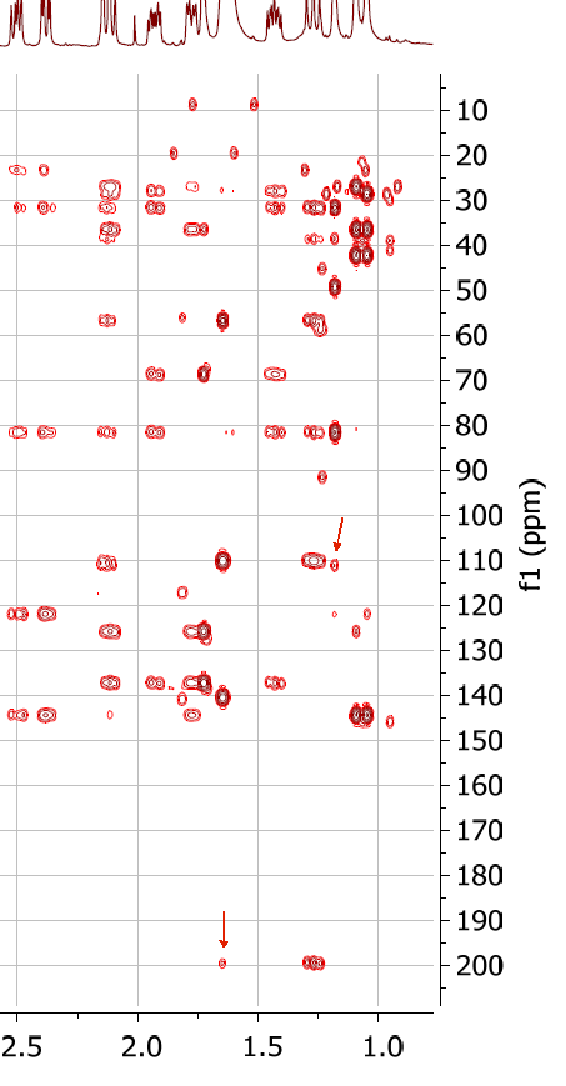
Figure 3. A part of the HMBC spectrum of compound 1.
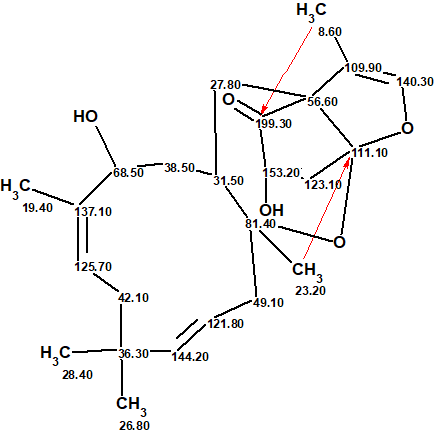
The structure of neosetophomone A with assigned 13C chemical shifts is shown below. This very unusual structure was unambiguously elucidated in a fully automatic mode in 5 seconds.
References
- T. El-Elimat, H. A. Raja, S. Ayers, S. J. Kurina, J. E. Burdette, Z. Mattes, R. Sabatelle, J. W. Bacon, A. H. Colby, M. W. Grinstaff, C. J. Pearce, N. H. Oberlies. (2019). Meroterpenoids from Neosetophoma sp.: A Dioxa[4.3.3]propellane Ring System, Potent Cytotoxicity, and Prolific Expression.Org. Lett. DOI: 10.1021/acs.orglett.8b03769
- M. E. Elyashberg, A. J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data. The Art of Solving Problems. Springer.


