June 1, 2019
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Nicotabin A
Nicotiana tabacum is the very well-known herbaceous plant of the Solenaceae family that provides the raw material for the tobacco industry. It is one of the very few plants that have been studied so extensively with as many as 2,500 compounds identified.
Feng et al [1] investigated the chemical constituents of N. tabacum leaves by a dereplication procedure, which resulted in the isolation of a novel compound, nicotabin A (1).
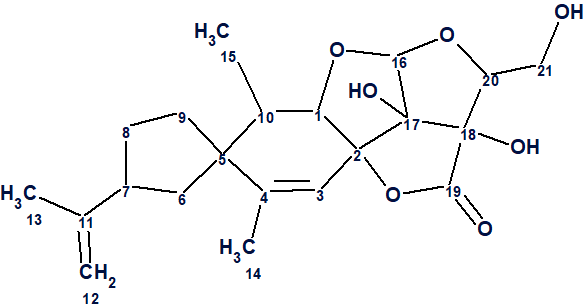
1
Its structure was elucidated by extensive spectroscopic methods, as well as by single-crystal X-ray diffraction. Compound 1 possessed a fused 5/6/5/5/5 ring system, representing a totally new carbon skeleton.
Nicotabin A (1) was isolated as colorless crystals from a MeOH solution. Its molecular formula C21H28O7 was determined from the positive ion HR-ESI mass spectrum, with a peak at m/z 415.1739 [M + Na]+ (calcd for C21H28O7Na: 415.1727), corresponding to eight degrees of unsaturation. The IR spectrum showed absorption bands for hydroxyl (3446 and 3361 cm−1), carbonyl (1774 cm−1), and olefinic (1647 cm−1) groups.
The spectroscopic data presented in [1] were used for challenging ACD/Structure Elucidator [2]. 1D and HSQC spectra were tabulated in [1], while only key HMBC and COSY correlations were given by arrows depicted in the structural formula of 1. The available spectroscopic data are shown in Table 1. There are three overlapping signals at 1.82 ppm and 2 at 1.71 ppm in the 1H spectrum, marked with red and blue on the table.
Table 1. Spectroscopic data of compound 1
| Label | δC | δCcalca | XHn | δH | M in 1H | COSY | H to C HMBC |
| C 1 | 88.800 | 85.690 | CH | 4.240 | u | 2.07 | C 2 |
| C 2 | 89.400 | 90.040 | C | ||||
| C 3 | 116.900 | 123.740 | CH | 5.210 | u | C 2 | |
| C 4 | 149.900 | 146.430 | C | ||||
| C 5 | 48.200 | 49.030 | C | ||||
| C 6 | 41.500 | 41.040 | CH2 | 1.490 | u | 2.51 | C 5 |
| C 6 | 41.500 | 41.040 | CH2 | 2.280 | u | ||
| C 7 | 47.100 | 46.390 | CH | 2.510 | u | 1.49, 1.52 |
C 11 |
| C 8 | 32.600 | 32.190 | CH2 | 1.520 | u | 1.71, 2.51 |
|
| C 8 | 32.600 | 32.190 | CH2 | 1.820 | u | ||
| C 9 | 35.500 | 33.590 | CH2 | 1.820 | u | C 5, C 4 |
|
| C 9 | 35.500 | 33.590 | CH2 | 1.710 | u | 1.52 | C 5, C 11 |
| C 10 | 40.700 | 44.580 | CH | 2.070 | u | 0.95, 4.24 |
C 5 |
| C 11 | 148.600 | 146.390 | C | ||||
| C 12 | 109.000 | 109.410 | CH2 | 4.660 | u | ||
| C 12 | 109.000 | 109.410 | CH2 | 4.710 | u | ||
| C 13 | 21.100 | 20.880 | CH3 | 1.710 | s | ||
| C 14 | 20.900 | 18.740 | CH3 | 1.820 | s | ||
| C 15 | 14.000 | 13.620 | CH3 | 0.950 | d | 2.07 | |
| C 16 | 111.500 | 114.710 | CH | 5.400 | u | C 20, C 1, C 17 |
|
| C 17 | 92.100 | 90.050 | C | ||||
| C 18 | 79.200 | 86.710 | C | ||||
| C 19 | 175.200 | 174.740 | C | ||||
| C 20 | 87.100 | 87.950 | CH | 4.000 | u | 3.61 | C 18 |
| C 21 | 61.700 | 60.910 | CH2 | 3.610 | u | 3.84, 4.00 |
|
| O 1 | OH | 3.840 | t | 3.61 | |||
| O 2 | OH | 5.050 | s | C 18, C 2, C 17 |
|||
| O 3 | OH | 5.730 | s | C 18, C 20, C 17, C 19 |
a 13C chemical shifts were predicted by the HOSE code based approach.
The molecular connectivity diagram (MCD) created automatically by the program is shown in Figure 1.
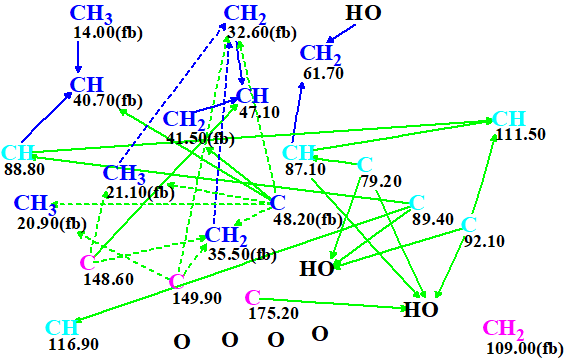
Figure 1. Molecular connectivity diagram.
MCD overview. The MCD contains ambiguous COSY (blue) and HMBC (green) correlations which are marked by dotted lines. These are a consequence of the presence in the 1H spectrum of overlapping signals at 1.82 and 1.71 ppm. There are seven light blue carbon atoms in the range of chemical shifts between 79.2 and 116.9 ppm. For these atoms, an ambiguous hybridization “not sp” (sp2 or sp3) was set by the program. In addition, no carbon atom has a label “ob” (connection to a heteroatom by a chemical bond is obligatory), and carbon CH2 109.00 has no correlations to other atoms (floating atom). The distinctive multiplicities in the 1H spectrum shown in Table 1 were introduced for the corresponding atoms displayed in the MCD. In spite of the fact that many atom properties are uncertain, and ambiguous HMBC and COSY correlations are present, no manual edits of the MCD were done. Structure generation accompanied with the 13C chemical shift prediction (the incremental approach was used [2]) and structural filtering was initiated.
Results: k = 28,070 → (filtering) → 489 → (removing duplicates) → 209, tg = 1 m 15 s. Additional 13C chemical shift prediction for the remaining structures using neural networks and HOSE code based methods was performed. The 1H chemical shifts were also predicted with the neural networks algorithm. Finally, the structures of the output file were ranked in descending order of average deviations of the 13C experimental chemical shifts from the predicted ones (denoted as dA),
calculated using the HOSE code based algorithm. The eight top structures of the ranked output file are shown in Figure 2.
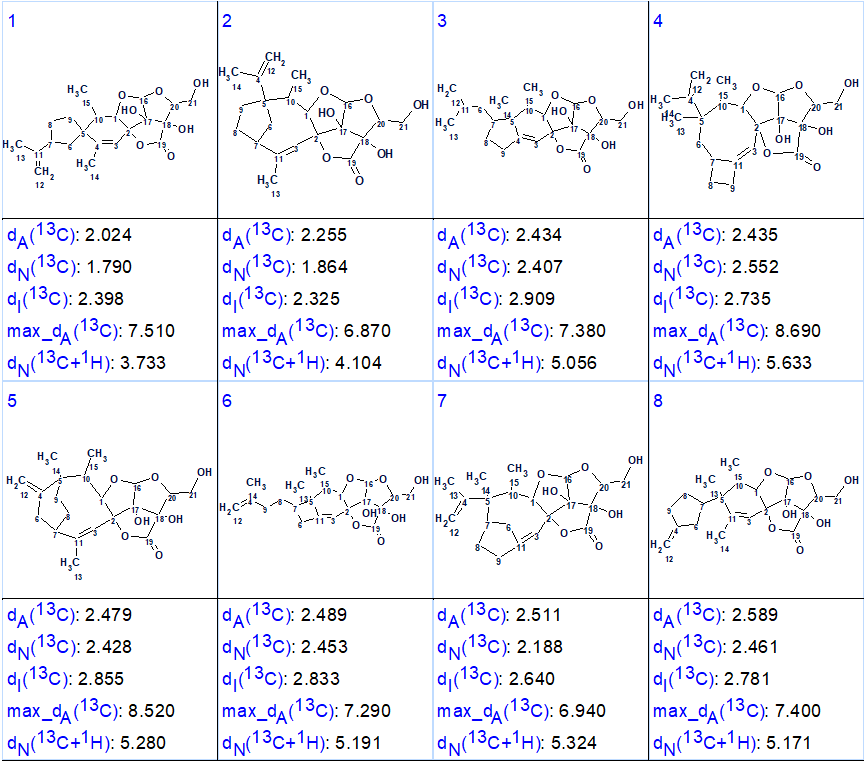
Figure 2. Eight top ranked structures of the ranked output file.
We see that the structure identified as best structure (#1) is identical to structure 1 which was determined for nicotabin A and confirmed by X-ray analysis in the article [1]. Thus, the solution to the problem was found in a fully automatic mode. It is worthy to note that all structures shown in Figure 2 contain the same large fragment in the right side of the molecule and all deviations are rather close. Nevertheless, the correct structure was selected by the program. Additional confidence to the structure could be gained if more NMR data were available. This would include additional HMBC correlations, e.g. to confirm the position of the CH2 at 109.0 ppm, and also some NOESY or ROESY data to narrow down the stereochemistry options. Because structure 1 contains 9 stereoisomers, its confirmation by DFT based chemical shift prediction [3,4] would probably be problematic and too time consuming. As a single crystal was available for the
compound 1, the authors [1] used X-ray crystallography for this goal instead.
It was interesting to see how the usage of multiplicities observed in the 1H NMR spectrum (Table 1, column “M in 1H”) influences the number of structures generated and the processing time. To test this, all the constraints arising from the 1H multiplicities were removed from the MCD and structure generation was repeated. Results: k =78,406 → (filtering) → 1,289 → (removing duplicates) → 566, tg = 3 m. The initial and final number of structures, as well as the generation time increased almost by a factor of three, while the top structures of the ranked file were the same. Therefore the utilization of 1H multiplicities accelerates the structure generation significantly, but one should be careful and realize that the introduction of at least one erroneous 1H multiplicity will lead to an inevitably erroneous solution.
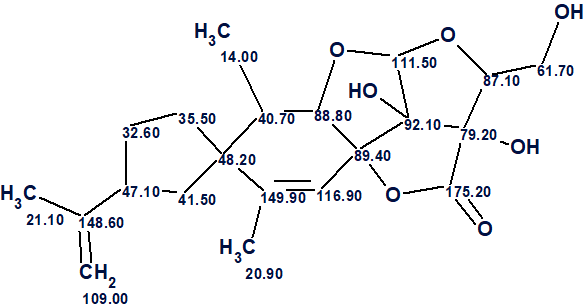
The structure of nicotabin A along with the 13C chemical shift assignment performed by ACD/Structure Elucidator is shown below.
References
- T. Feng, X.-M. Li, J. He, H.-L. Ai, H.-P. Chen, X.-N. Li, Z.-H. Li, J.-K. Liu. (2017). Nicotabin A, a Sesquiterpenoid Derivative from Nicotiana tabacum. Org. Lett., 19: 5201−5203.
- M. E. Elyashberg, A. J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data. The Art of Solving Problems. Springer.
- A. V. Buevich, M. E. Elyashberg. (2016) Synergistic combination of CASE algorithms and DFT chemical shift predictions: a powerful approach for structure elucidation, verification and revision. J. Nat. Prod., 79 (12): 3105–3116.
- A.V. Buevich, M. E. Elyashberg. (2018). Towards unbiased and more versatile NMR-based structure elucidation: A powerful combination of CASE algorithms and DFT calculations. Magn. Reson. Chem., 56: 493–504. DOI: 10.1002/mrc.4645


