March 1, 2017
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Phomopsterone A
Steroids are arguably one of the most important groups of small molecules in biology. They are also the second largest drug category in the market. Several steroid-based drugs are being used for the therapy of a wide variety of diseases primarily because of their anti-inflammatory action. As a result chemists and biologists have an ongoing interest in their chemical structure and biological properties.
The terrestrial and marine-derived fungi that belong to the genus Phomopsis are known for their potent biosynthetic capabilities producing structurally diverse secondary metabolites. The fungus Phomopsis sp. TJ507A, which was originally isolated from the medicinally important plant Phyllanthus glaucus, was recently analyzed by Hu and co-workers [1]. Two functionalized ergostane-type steroids, phomopsterones A (1) and B were isolated. Phomopsterone A was chosen for challenging ACD/Structure Elucidator.
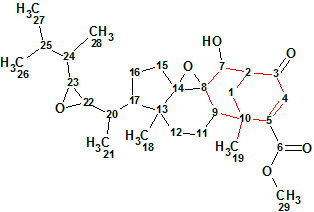
1
Structurally phomopsterone A (1) is a highly oxygenated and unique rearranged ergostane-type steroid featuring a unique bicyclo[3.3.1]nonane motif (colored in red) with an α- oriented Me-19 group. Its structure was confirmed by single-crystal X-ray analysis [1].
Phomopsterone A was obtained as colorless crystals with the molecular formula C29H42O6 as concluded from its 13C NMR and HRESIMS ([M + Na]+ m/z 509.2879, calcd for 509.2879) data corresponding to nine sites of unsaturation. The IR spectrum of 1 revealed the presence of hydroxyl (3500 cm−1) and carbonyl (1728, ester, and 1668 cm−1, conjugated ketone) groups.
To elucidate the structure, authors [1] used 1D and 2D NMR data, including HSQC, HMBC and COSY. COSY and some key HMBC correlations were shown graphically in the article [1]. The available NMR spectroscopic data (Table 1) and the molecular formula of 1 were entered into ACD/Structure Elucidator.
Table 1. Phomopsterone A. NMR spectroscopic data
| Label | δC | δC Calc | XHn | δH | COSY | H to C HMBC |
| C 1 | 39.4 | 39.56 | CH2 | 2.1 | 2.75 | C 3 |
| C 1 | 39.4 | 39.56 | CH2 | 2.32 | ||
| C 2 | 51.1 | 49.39 | CH | 2.75 | 2.10, 3.51 |
C 3 |
| C 3 | 199.7 | 202.03 | C | |||
| C 4 | 134.4 | 131.76 | CH | 6.59 | C 2, C 5, C 6, C 10 | |
| C 5 | 153.9 | 156.63 | C | |||
| C 6 | 167.4 | 166.31 | C | |||
| C 7 | 71.5 | 72.11 | CH | 3.51 | 2.75 | C 8, C 9 |
| C 8 | 64.5 | 66.24 | C | |||
| C 9 | 43.8 | 44.47 | CH | 2.13 | ||
| C 10 | 38.4 | 39.68 | C | |||
| C 11 | 18.6 | 19.6 | CH2 | 1.34 | 1.07 | |
| C 11 | 18.6 | 19.6 | CH2 | 1.63 | ||
| C 12 | 38.3 | 34.15 | CH2 | 1.07 | 1.34 | |
| C 12 | 38.3 | 34.15 | CH2 | 1.7 | ||
| C 13 | 41.2 | 42.56 | C | |||
| C 14 | 71.3 | 77.56 | C | |||
| C 15 | 26.9 | 27.09 | CH2 | 1.96 | ||
| C 15 | 26.9 | 27.09 | CH2 | 1.65 | 1.57 | C 8, C 13, C 14 |
| C 16 | 26.8 | 25.9 | CH2 | 1.95 | ||
| C 16 | 26.8 | 25.9 | CH2 | 1.57 | 1.54, 1.65 | |
| C 17 | 56.9 | 48.25 | CH | 1.54 | 1.25, 1.57 | |
| C 18 | 15.5 | 22.2 | CH3 | 0.79 | C 12, C 13, C 14, C 17 | |
| C 19 | 24.7 | 25.71 | CH3 | 1.36 | C 1, C 5, C 9, C 10 | |
| C 20 | 38.5 | 39.79 | CH | 1.25 | 1.00, 1.54, 2.54 | |
| C 21 | 16.8 | 16.38 | CH3 | 1 | 1.25 | |
| C 22 | 64.2 | 64.6 | CH | 2.54 | 1.25, 2.43 | |
| C 23 | 61.2 | 62.28 | CH | 2.43 | 1.03, 2.54 | |
| C 24 | 42.4 | 42.12 | CH | 1.03 | 0.94, 1.64, 2.43 | |
| C 25 | 31.3 | 31.34 | CH | 1.64 | 0.90, 0.92, 1.03 | |
| C 26 | 20.7 | 20.59 | CH3 | 0.92 | 1.64 | |
| C 27 | 19.7 | 18.99 | CH3 | 0.9 | 1.64 | |
| C 28 | 13.7 | 12.96 | CH3 | 0.94 | 1.03 | |
| C 29 | 52.5 | 52.43 | CH3 | 3.75 | C 6 |
A Molecular Connectivity Diagram (MCD) automatically produced by the software is shown in Figure 1.
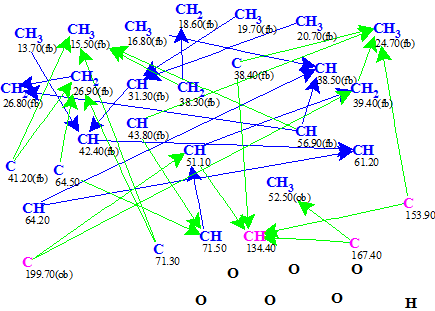
Figure 1. Phomopsterone A. Molecular Connectivity Diagram.
MCD overview. Hybridizations of all carbon atoms were automatically set and marked by the software. The sp3 hybridization is denoted by blue color and sp2 is pink. Labels “fb” and “ob” are used to distinguish atoms for which connection to a heteroatom (oxygen in this case) are either forbidden or obligatory. HMBC connectivities are shown as green arrows and COSY as blue ones. No user edits of the MCD were made.
As there were no contradictions found in 2D NMR data, a Strict Structure Generation was initiated which gave the following results: k = 30→16→16, tg = 1 s.
Following the general methodology [2] 13C chemical shifts were predicted by three methods (Incremental, Neural Networks, HOSE code-based). The resulting output structural file was ranked in increasing order of the average deviation dA (13C chemical shifts were calculated by HOSE code based approach). The six top structures of the ranked file are presented in Figure 2.
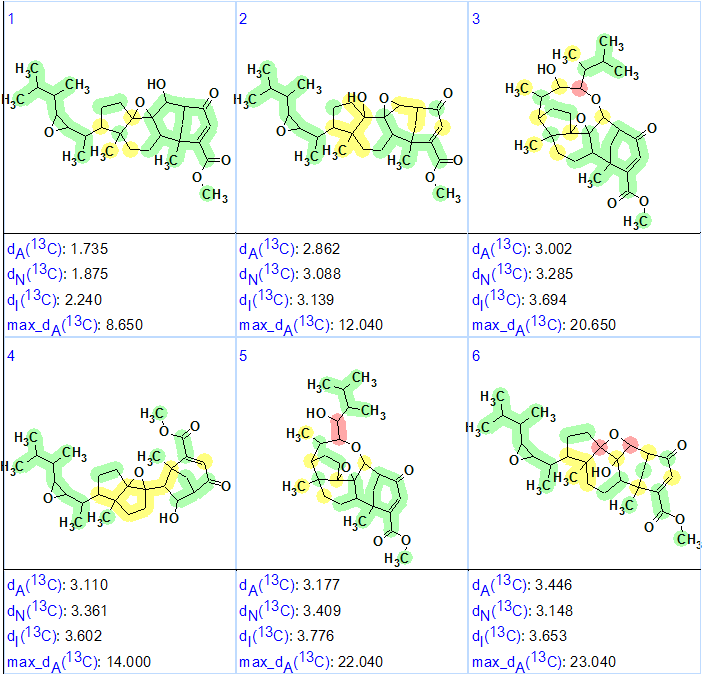
Figure 2. The six top structures of the ranked file for Phomopsterone A. The color attributes of the 13C chemical shift prediction accuracy are: ±3 ppm – green, between 3 and 15 ppm – yellow, greater than 15 ppm – red.
Figure 2 shows that the top-ranked structure #1 coincides with the structure of Phomopsterone A determined in the original article and confirmed by single crystal x-ray diffraction [1]. The deviations calculated as well as the accuracy of the carbon atom chemical shift prediction marked with color allowed us to positively conclude that the solution obtained using ACD/Structure Elucidator Suite is correct.
The elucidated structure with automatically assigned 13C chemical shifts is shown below
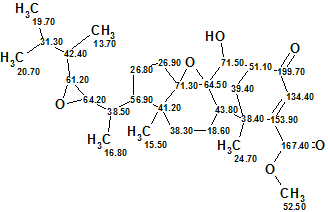
In summary, a new complex natural compound containing a unique structural motif was fully, automatically and confidently identified by ACD/Structure Elucidator in one second.
References
- Z. Hu, Y. Wu, S. Xie, W. Sun, Y. Guo, X.-N. Li, J. Liu, H. Li, J. Wang, Z. Luo, Y. Xue, Y. Zhand. (2017). Phomopsterones A and B, Two Functionalized Ergostane-Type Steroids from the Endophytic Fungus Phomopsis sp. TJ507A. Org. Lett. 19(1): 258–261.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.


