February 1, 2012
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Phoriospongin A
The Initial Challenge
In 2002, Robert Capon and co-workers1 identified two new nematocidal depsipeptides, identified as phoriospongins A and B through bioassay-directed fractionation of two southern Australian sponges, Phoriospongia sp. and Callyspongia bilamellata. The structures of the phoriospongins were determined by detailed spectroscopic analysis and comparison with the previously reported sponge depsipeptide cyclolithistide A. The initial experimental data was then submitted to the Structure Elucidator Challenge to determine if the software could propose the same structure.
The Evidence
- The molecular formula of phoriospongin A was determined to be C52H82N11O15Cl
- The following data were acquired:
- 2D 1H-1H COSY (59 correlations), 1H-13C HMBC (71 correlations)
The CASE (Computer Assisted Structure Elucidation) Investigation
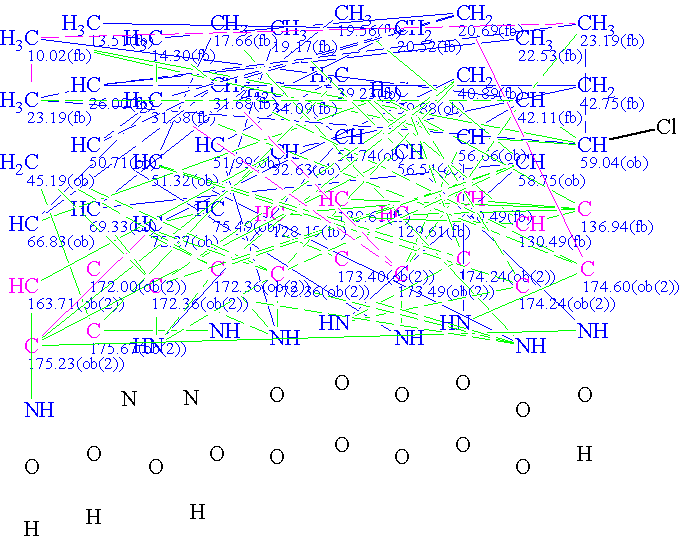
The Molecular Connectivity Diagram produced by Structure Elucidator
-
- ACD/Structure Elucidator produced a Molecular Connectivity Diagram (MCD) from the experimental data.
- A built-in algorithm, specifically designed to check the consistency of all the experimental data, reported non-standard connectivities (NSCs) within the data.
- The program automatically detected and improved five of the six NSCs—their lengths were increased by one bond (the lengths were set from 1 to 2 skeletal bonds for COSY and from 1 to 3 skeletal bonds for HMBC). The sixth NSC was corrected by hand assuming that the corresponding 2D NMR peak is weak.
- The program generated 624 structures over a period of 17 hours; the long generation time is due to a) the large size of the molecule, and b) the presence of six long-range NSCs, which dramatically increases number of structures fitting the set of 2D NMR data.
- Structure 1 ranked in the first position based on the lower dA value; the average deviation (dA) is a comparison between the experimental and predicted 13C chemical shifts calculated by HOSE code based approach.
| Structure #1 | Structure #2 | Structure #3 |
| dA = 1.132 | dA = 1.347 | dA = 1.376 |
| dF = 2.316 | dF = 2.255 | dF = 2.373 |
| dH = 0.294 | dH = 0.306 | dH = 0.317 |
| dΣ = 4.074 | dΣ = 4.403 | dΣ = 4.549 |
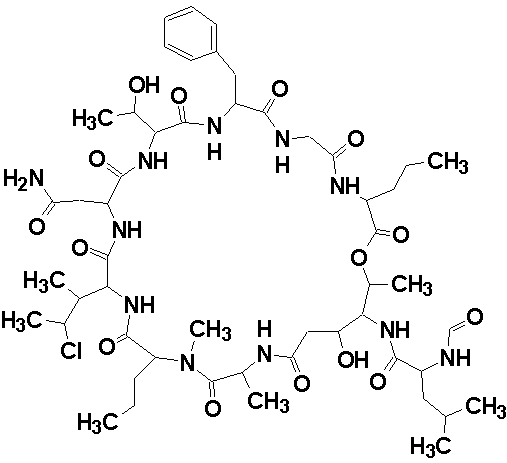
Structure 1
- Structure 1 matches the structure published by the original authors who solved it by traditional means.
Conclusions
For this example, ACD/Structure Elucidator detected and improved all but one of the non-standard COSY and HMBC correlations, and was able to successfully determine the correct structure for a large cyclic peptide comprising of ~80 skeletal atoms.
- dF—the average deviation between the experimentally observed carbon chemical shifts and the carbon chemical shifts predicted using fast (increment based) prediction methods
- dH—the average deviation between the experimentally observed proton chemical shifts and the predicted proton chemical shifts using accurate (HOSE code based) prediction methods
- dΣ = dA + 20-dH—generalized match factor that takes into account deviations calculated by the accurate prediction methods both for 13C and 1H NMR spectra
References
- Robert J. Capon, Joanne Ford, Ernest Lacey, Jennifer H. Gill, Kirstin Heiland, and Thomas Friedel J. Nat. Prod., 65:358–363, 2002.


