November 1, 2013
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Phosphoiodyn A
Polyacetylenic natural products, characterized by carbon-carbon triple bonds or alkynyl functional groups, are widely distributed in natural objects. Polyacetylenes vary by a number of structural features, including the number of triple bonds, chain lengths, and the substituted functional groups. A number of polyacetylenes have been isolated from marine sponges.
They exhibit diverse bioactivities such as cytotoxic, antiviral, antifouling, RNA-cleaving, and enzyme-inhibitory activities.
Kim et al [1] isolated and identified two unprecedented phosphorus-containing iodinated polyacetylenes, Phosphoiodyn A and B, from a Korean marine sponge Placospongia sp. Here we will describe the structural elucidation of Phosphoiodyn A (1).
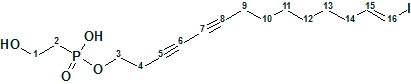
1
Phosphoiodyn A (1) was obtained as an amorphous solid. According to the authors [1], the molecular formula of 1 was deduced as C16H24O4PI based on the pseudomolecular ion peak at m/z 438.0459 [M+H]+ in the HR-FABMS and 13C NMR data. But when the chemical composition CHOPI was postulated for the unknown and the value of the pseudomolecular ion peak was input in the Molecular Formula Generator of ACD/Structure Elucidator Suite, the following result was obtained (Figure 1):
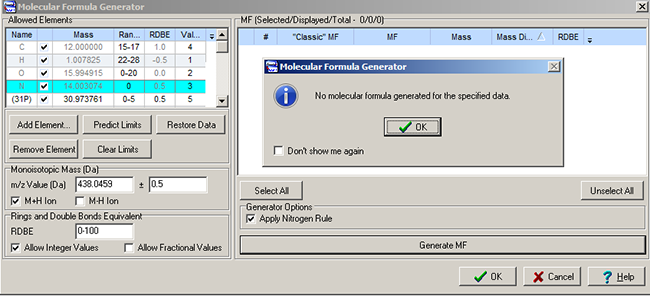
Figure 1. Phosphoiodyn A. The result of molecular formula generation at [M+H]+ 438.0459 when chemical composition was assumed to be CHOPI. No molecular formula has been generated.
The result is not unexpected: if a molecule does not contain a nitrogen atom the value of the pseudomolecular ion peak can not be even, therefore [M+H]+ 439.0459 should be input into the Molecular Formula Generator. Under this condition, the molecular formula C16H24O4PI was generated (Δm=0.0076). 1H, 13C, HSQC, COSY and HMBC spectroscopic data of compound 1 are presented in Table 1. The 1H NMR spectrum of 1 displayed two methines and ten methylenes, two of which, C1 and C3, authors [1] assigned as oxygenated methylenes. Running a few steps forward, it is worthwhile to place attention on the discrepancy between experimental (35.5) and predicted (55.5) values of chemical shifts assigned to carbon atom C1 (Table 1). The 13C NMR spectrum, in combination with HSQC and HMBC spectra, indicated four fully substituted carbons at dC 73.1, 66.1, 64.8, and 77.1 which allowed authors the assignment of a diyne unit.
Table 1. Phosphoiodyn A. Spectroscopic NMR data (click to expand)
| Label | δC | δC calc | CHn | δH | M(J) | COSY | C HMBC |
| C 1 | 35.5 | 54.98 | CH2 | 3.14 | u | 1.89 | C 2 |
| C 2 | 24.5 | 30.36 | CH2 | 1.89 | u | 3.14 | C 1 |
| C 3 | 61.9 | 63.03 | CH2 | 3.94 | u | 2.6 | C 4, C 5 |
| C 4 | 21.2 | 21.77 | CH2 | 2.6 | u | 3.94 | C 5, C 6, C 3, C 7 |
| C 5 | 73.1 | 74.61 | C | ||||
| C 6 | 66.1 | 66.27 | C | ||||
| C 7 | 64.8 | 64.78 | C | ||||
| C 8 | 77.1 | 77.29 | C | ||||
| C 9 | 18.2 | 19.1 | CH2 | 2.25 | u | 1.5 | C 6, C 10, C 8 |
| C 10 | 27.9 | 28.1 | CH2 | 1.5 | u | 2.25, 1.39 | C 11, C 9, C 8 |
| C 11 | 28 | 27.94 | CH2 | 1.39 | u | 1.5 | C 10, C 12 |
| C 12 | 28.2 | 27.92 | CH2 | 1.32 | u | 1.43 | C 13, C 11 |
| C 13 | 28 | 29.1 | CH2 | 1.43 | u | 2.07, 1.32 | C 15, C 14 |
| C 14 | 35.5 | 36 | CH2 | 2.07 | u | 1.43, 6.52 | C 16, C 15, C 13 |
| C 15 | 147 | 146.3 | CH | 6.52 | u | 6.10, 2.07 | C 13, C 14, C 16 |
| C 16 | 73.6 | 74.5 | CH | 6.1 | u | 6.52 | C 15, C 14 |
The Molecular Connectivity Diagram (MCD) modified in accord with some of the authors’ assumptions regarding atom properties is presented in Figure 2.
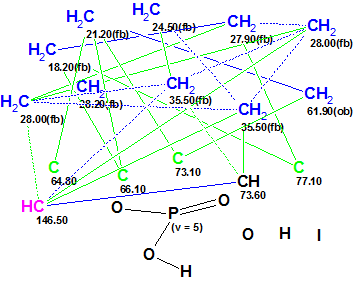
Figure 2. Phosphoiodyn A. Molecular Connectivity Diagram. Four sp-hybridized carbon atoms are marked by green color.
MCD overview. The MCD was generated with the check box “Allow sp Carbons” selected in the Create MCDs Options window, because the presence of triple bonds between carbon atoms was postulated based on the sample origin and chemical knowledge. As a result, carbon atoms C 64.80 – C 77.10 were marked by the program with the attribute of hybridization “not defined” (and the atoms originally colored in black). Following the authors’ assumptions, atoms C 64.8, C 66.1, C 73.1 and C 77.1 were assigned by the user as sp-hybridized (Edit Properties of Atom #) and the program colored them in characteristic green color. Properties of the atom CH(73.60, 6.10) were left without any change, as its hybridization might be either sp2 or sp3. The methylene 63.9 received a label “ob“, while all remaining methylene groups (including CH2 35.5) were labeled by the program with a label “fb” on the basis of the Atom Property Correlation Table (APCT). Finally, the phosphorus-containing fragment was drawn in the MCD by hand, as was suggested by authors.
An attempt at structure generation immediately gave an empty structural file i.e. no structures were able to be generated. It was proposed that the next attempt be performed with the APCT switched off (“not used” is selected) considering the unknown structure is unprecedented. This second attempt was performed in Fuzzy Structure Generation mode, with “Set Options Automatically” selected, producing the result: k=104→104→52, tg = 0.2 s. Connectivity lengths were elongated by one chemical bond (a=1) at m=1. Structural file ranking delivered the best structures with an average deviation range of 10-12 ppm, which can be considered as indication to increase m from 1 to 2. The third run gave the following result: k=48→48→12, tg = 1.2 s, and Figure 3 shows the top structures of the ranked output file.
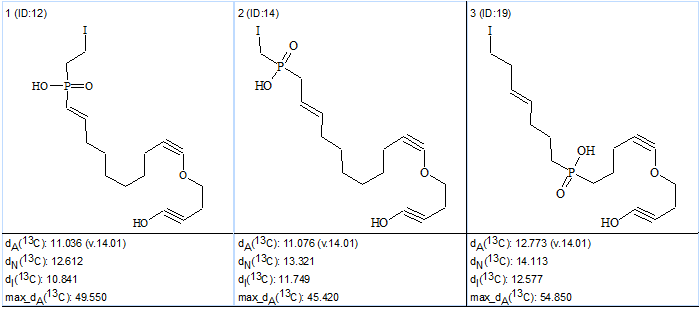
Figure 3. Phosphoiodyn A. Top structures of the ranked output file at m=2.
Figure 3 shows that both average and maximum deviations are extremely high, which can be considered as a hint that the initial set of axioms may need to be revised. As shown in Figure 2, two carbons with overlapped signals in the 13C spectrum at 35.5 ppm are labeled as carbons for which neighboring heteroatoms are forbidden. Meanwhile, one of these atoms, C1, is a neighbor of a hydroxyl group in structure 1 according to the authors’ [1] suggestion. Therefore, to generate structure 1 and retain it during spectral filtering, it is necessary to remove the label “fb” at atoms C 35.5 (select “not defined” in the Edit Properties of Atom # dialog window) and switch off both APCT (choose “not used” in the CSB Generator Options dialog window) and Filter. Under these conditions, the results of Fuzzy Structure Generation were: k=101→101→20, tg = 1 s, 2 from 26 connectivities having been extended during generation, with 94 out of 325 possible connectivity combinations used during generation. The three top structures of the ranked output file are presented in Figure 4.
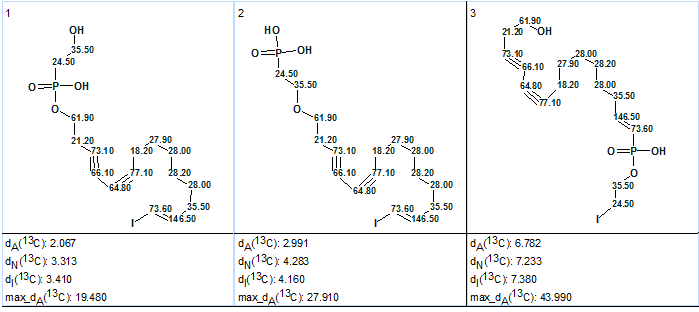
Figure 4. Phosphoiodyn A. Top structures of the ranked output file. Fuzzy Structure Generation (m=2) under conditions that an oxygen atom may be a neighbor of C 35.5.
We see that the first ranked structure does coincide with the authors’ structural suggestion [1], and average deviations are acceptable if we take into account that the structure is unprecedented. However the maximum deviation (19.5 ppm) is too high. The protocol of 13C chemical shift prediction for –CH2-OH is presented in Figure 5.
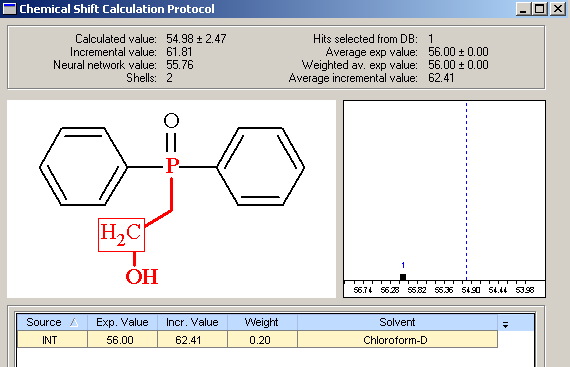
Figure 5. Phosphoiodyn A. Chemical shift calculation protocol generated for the carbon CH2-OH of the structure #1, Figure 4.
Figure 5 shows that only one hit was found in the database for 13C chemical shift calculation and the predicted value is 55 ppm; this means the difference between the experimental and calculated values is ~20 ppm. On the other hand, as seen from Table 1, the predictions for other carbon atoms are excellent. In addition, the following two reference structures were found in the ACD/CNMR database:
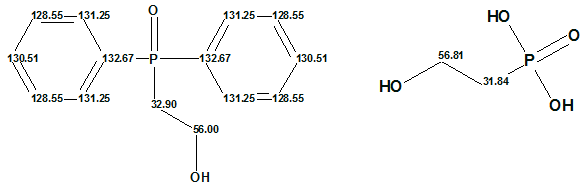
Figure 6: Structures identified in the ACD/CNMR Database as references for the predicted chemical shifts of the unknown structure.
Thus, we can conclude that the chemical shift 35.5 ppm can not be assigned to the methylene group CH2-OH, and therefore either the published experimental data or structure 1 are erroneous.
The results obtained with Structure Elucidator were sent to the authors [1]. As a result of compound reinvestigation the authors revised structures both of Phosphoiodyn A and B (structures are very similar [2]). It turned out that the reason for the wrong initial structure was a mistaken assumption of the unknown compound’s possible chemical composition. The “axiom” on the chemical composition of the unknowns was revised by the authors: the possibility of the presence of a nitrogen atom was admitted. With the new axiom (chemical composition CHNOPI) and the pseudomolecular ion peak at m/z [M+H]+ 438.0692, the molecular formula C16H25O3NPI was generated and reliably selected as the most probable one with Dm=0.0003. The revised molecular formula was input into the program and new Molecular Connectivity Diagram was created. In contrast to the previous MCD (Figure 2), no labels specifying the possibility of CH2 35.5 carbons to have neighboring heteroatoms were assigned. This is because the program took into account the fact that a nitrogen atom is present in the molecular formula (i.e. presence of a fragment CH2-N is allowed). Fuzzy Structure Generation with parameters used during the last program run (m=2, a=1) gave the following result: k=101→ 20, tg = 1 s. Three top structures of the ranked output file are shown in Figure 7.
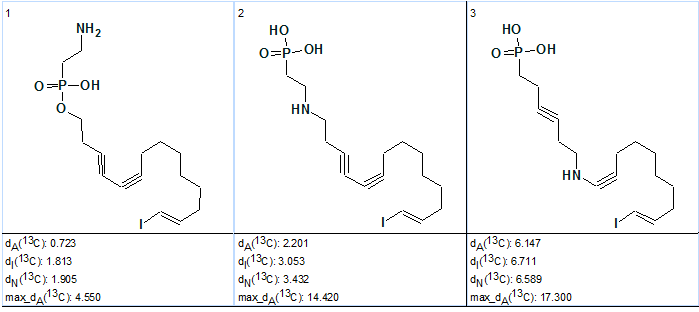
Figure 7. Phosphoiodyn A. Top structures of the ranked output file generated from the molecular formula C16H25O3NPI.
Figure 7 shows that the best structure is characterised by small average and maximum deviations and it can be reliably selected as the real structure of Phosphoiodyn A (2) which coincides with the revised structure [2].
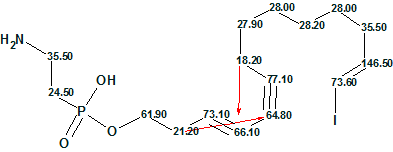
2
Here red arrows denote two 4JCH HMBC correlations. It is worthy to note that only two structures (#1 and #2 in Figure 7) were generated in 0.2 s when the Fuzzy Structure Generation was repeated with APCT and Filter switched on.
This example demonstrates how a wrong assumption (axiom) about the conceivable chemical composition leads to inferring an erroneous structure. It also underlines that chemical shift prediction for a structural hypothesis allows one to deduce that the proposed structure is erroneous, and that the interpretations of experimental data need to be re-checked and/or revised.
References
- Phosphoiodyns A and B, Unique Phosphorus-Containing Iodinated Polyacetylenes from a Korean Sponge Placospongia sp, H. Kim, J. Chin, H. Choi, K. Baek, T.-G. Lee, S. E. Park, W. Wang, D. Hahn, I.Yang, J. Lee, B. Mun, M. Ekins, S.-J. Nam, H. Kang. Org. Lett., 15(1):100–103, 2013.
- Phosphoiodyns A and B, Unique Phosphorus-Containing Iodinated Polyacetylenes from a Korean Sponge Placospongia sp Additions and Corrections, H. Kim, J. Chin, H. Choi, K. Baek, T.-G. Lee, S. E. Park, W. Wang, D. Hahn, I.Yang, J. Lee, B. Mun, M. Ekins, S.-J. Nam, H. Kang. Org. Lett. Volume 1, 2013.


