July 1, 2013
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Polypropionat
Marine mollusks of the genus Siphonaria, commonly known as false limpets, are shelled, air-breathing herbivores that are believed to have a marine ancestry. When disturbed, siphonariid limpets secrete a sticky white mucus from their lateral pedal glands. The mucus released by Siphonaria species is rich in polypropionate secondary metabolites. Colonies of S. oculus occur along the southern and eastern coast of South Africa and southern Mozambique. The S. oculus specimens were steeped in acetone, and the acetone extract was subjected to extensive chromatography to afford three new polypropionate metabolites. Spectroscopic data of Polypropionate 1 are used here to describe the structure elucidation of this compound using CASE techniques.
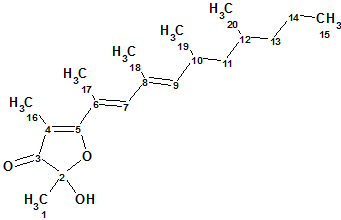
1
Polypropionate 1 was isolated as a colorless oil, with a molecular formula established as C20H32O3 from HREIMS data [m/z 320.2350 [M+] (calculated for C20H32O3, 320.2351)], which implied five degrees of unsaturation. The characteristic bands at 3339 and 1686 cm-1 in the IR spectrum of 1 indicated the presence of hydroxy and carbonyl functionalities, respectively.
1H, 13C, HSQC and HMBC NMR data tabulated in the article [1] are presented in Table 1.
Table 1. Spectroscopic NMR data.
| Label | δC | δC calc | CHn | dH | M(J) | C HMBC |
| C 1 | 22.2 | 21.93 | CH3 | 1.55 | S | C 2, C 3, C 16 |
| C 2 | 102 | 102.15 | C | |||
| C 3 | 202.8 | 203.97 | C | |||
| C 4 | 108.5 | 108.42 | C | |||
| C 5 | 184.8 | 182.99 | C | |||
| C 6 | 122.8 | 125.8 | C | |||
| C 7 | 139.9 | 141.24 | CH | 6.26 | d(1.2)* | C 17, C 18, C 9, C 5 |
| C 8 | 130.7 | 131.24 | C | |||
| C 9 | 141.5 | 144.02 | CH | 5.2 | d(9.8) | C 18, C 7, C 11 |
| C 10 | 30.5 | 32.19 | CH | 2.49 | u | C 9, C 19, C 11, C 8 |
| C 11 | 45.2 | 45.21 | CH2 | 1.05 | u | C 13, C 20, C 19 |
| C 11 | 45.2 | 45.21 | CH2 | 1.21 | u | C 12, C 10, C 9 |
| C 12 | 30.6 | 30.54 | CH | 1.28 | u | C 13, C 11, C 10 |
| C 13 | 40.1 | 39.46 | CH2 | 1.06 | u | C 20 |
| C 13 | 40.1 | 39.46 | CH2 | 1.19 | u | C 15, C 11, C 12 |
| C 14 | 20.1 | 20.19 | CH2 | 1.29 | u | C 13, C 12, C 15 |
| C 14 | 20.1 | 20.19 | CH2 | 1.23 | u | |
| C 15 | 14.5 | 14.17 | CH3 | 0.85 | t(7.2) | C 14, C 13 |
| C 16 | 6.3 | 6.99 | CH3 | 1.52 | S | C 6, C 4, C 5, C 3, C 7 |
| C 17 | 21.8 | 15.19 | CH3 | 1.98 | S | C 5, C 16, C 7, C 6, C 4, C 8, C 18 |
| C 18 | 15.1 | 15.63 | CH3 | 1.69 | S | C 8, C 7, C 11, C 19, C 10, C 9 |
| C 19 | 21 | 21.54 | CH3 | 0.87 | d(6.5) | C 12, C 10, C 11, C 9 |
*Small coupling constant can be interpreted as an indication to the absence of hydrogens attached to the neighbor skeleton atoms.
The Molecular Connectivity Diagram (MCD) created from the NMR data is shown in Figure 1.
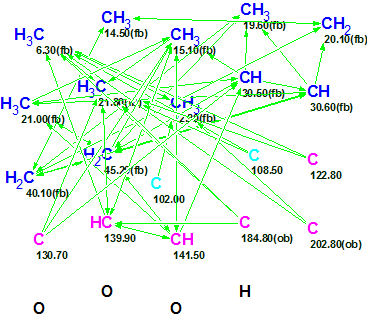
Figure 1. Molecular Connectivity Diagram of Polypropionate 1.
MCD overview. Two light blue atoms C 102.00 and C 108.5 can exist in the molecule either in sp2-hybridized state or in sp3 if formation of the O-C-O substructure is possible. To demonstrate the results of automatic problem solving with ACD/Structure Elucidator, no edits were made to the MCD. Numbers of hydrogen atoms attached to the neighbor carbons were input in accordance with 1H multiplicities shown in column M(J) of Table 1.
MCD checking by the software revealed the presence of at least 4 HMBC nonstandard correlations (NSC). Therefore the Fuzzy Structure Generation was initiated with the options m=4-20, a=16, “Stop Generation when Structures Generated”. These options indicate that the expected number of nonstandard connectivities may be between 4 and 20, and the length of NSCs is unrestricted. 13C chemical shift prediction was also carried out during the structure generation to reject structures characterized by average 13C chemical shift deviations d>4 ppm. Results: k=6→2→1, tg = 56 m, 12 from 43 connectivities have been extended and 18,440,500 from 15,338,678,264 (0.1 %!) possible connectivity combinations were used.
The single output structure (dA=1.12, dN=1.10, dI=1.45 ppm) coincided with the structure of Polypropionate 1. 13C chemical shift assignment, as well as all nonstandard connectivities are displayed on structure 1a:
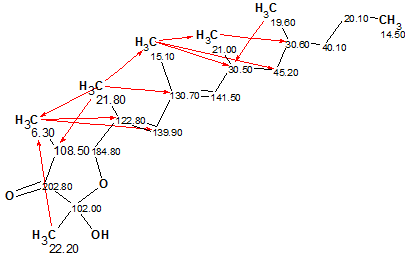
1a
Some interesting observations can be made when analyzing the solution obtained.
i) The number of NSCs in the HMBC data turned out to be 12, and four of nonstandard connectivities were 4 C-C chemical bonds length (5JCH) as shown in Figure 2, taken from the original Supporting Information [1]. Note that peaks corresponding to these correlations are of intensity comparable with that common for correlations of standard length (2-3JCH).
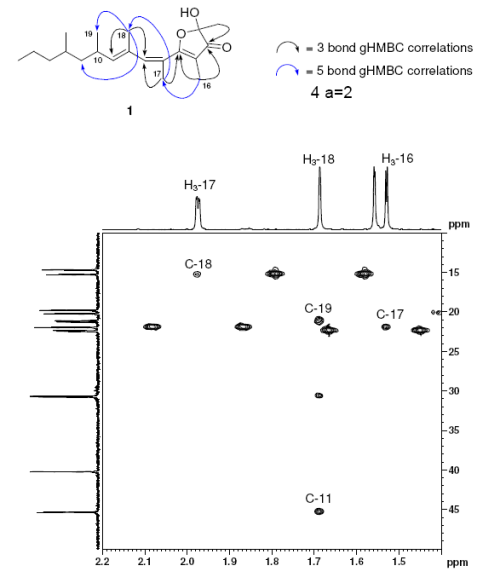
Figure 2. Polypropionate 1. A region (F1 = δ 1.4–2.2 ppm; F2 = δ 10–50 ppm) of the gHMBC spectrum
(CDCl3, 600 MHz) of 1 illustrating the five bond gHMBC correlations. The accompanying structure shows the key gHMBC correlations used to elucidate the structure of 1.
ii) Only 0.1% of the total number of all possible connectivity combinations were used in the structure generation process, due to the algorithm that is capable of selecting the most relevant connectivity combinations. Nevertheless the processor time consumed during the structure generation was about an hour. We can expect that trying ~15 billion possible combinations would need for about 40 days of processor time, i.e. the solution of the problem would become practically impossible.
iii) In spite of the high number of nonstandard correlations having different lengths, the unambiguous solution to the problem was found completely automatically.
iiii) The most advanced mode of the Fuzzy Structure Generation, where no restriction is imposed on the lengths of nonstandard connectivities (a = 16), allowed us to solve the problem under condition that HOSE data contained 8 NSCs of 4JCH type and 4 NSCs of 5JCH type. Note that real number of NSCs and their lengths became known only when the problem has been solved.
The problem was solved but the Fuzzy Structure Generation consumed about an hour of processor time. We could expect that application of the Fragment Library could help to reduce this time. For this goal a fragment search using 13C chemical shifts was performed in the Fragment Library which was completed with detection of 73 fragments whose sub-spectra are “projected” on the experimental 13C NMR spectrum.
Then the command Structure Elucidation/Create MCD Using Fragments was initiated with the Options presented in Figure 3. All options influencing the MCD creation were set in such a way that all procedures were automatically executed. As a result only one MCD was created in which one found fragment was “inplanted” (Figure 4).
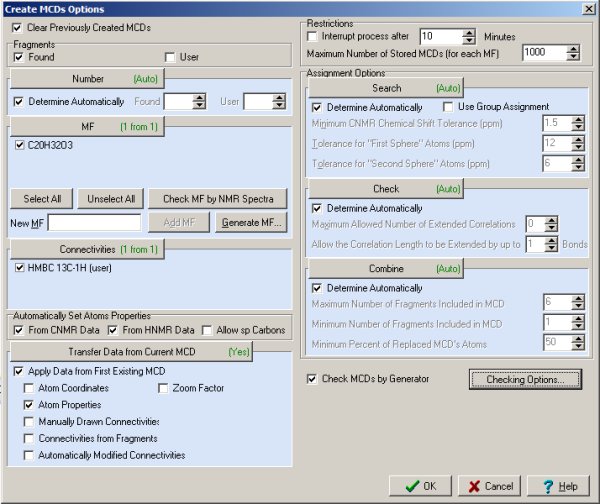
Figure 3. Polypropionate 1. Options of MCD creation.
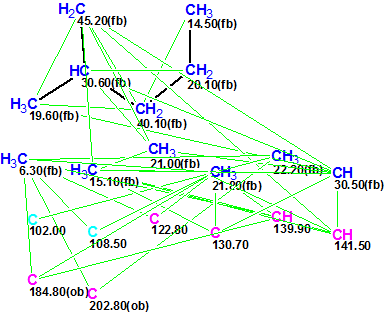
Figure 4. Polypropionate 1. Molecular Connectivity Diagram containing one Found Fragment.
The Fuzzy Structure Generation was initiated again with options shown in Figure 5.
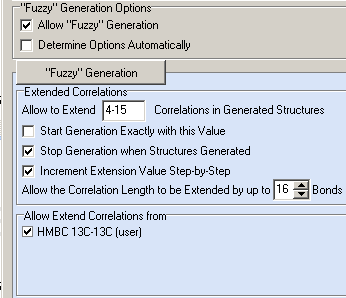
Figure 5. Polypropionate 1. Options of the Fuzzy Structure Generation from the Molecular Connectivity Diagram created using Found Fragments.
The following results were obtained: k=6→6→3, tg=52 s , 12 from 33 connectivities have been extended during generation, 500,900 from 354,817,320 (0.1%) possible connectivity combinations have been using during generation. The ranked output file is presented in Figure 6.
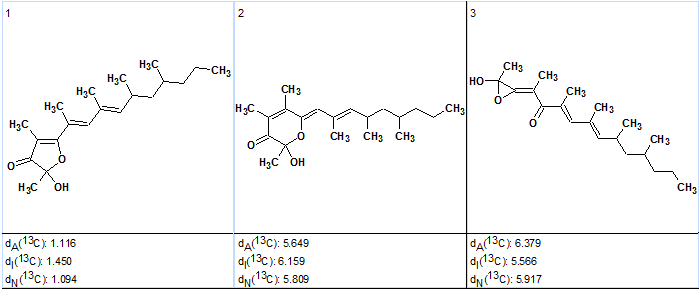
Figure 6. Polypropionate 1. Ranked output file.
As Figure 6 shows the correct structure was ranked first and the competing strictures can be rejected with high reliability due to large difference between deviations. At the same time, computational expenses reduced from 56 m down to 52 s, i.e. by more than a factor of 60.
References
- C. L. Bromley, W. L. Popplewell, S. C. Pinchuck, A. N. Hodgson, M. T. Davies-Coleman, Polypropionates from the South African Marine Mollusk Siphonaria oculus J. Nat. Prod., 2012, 75 (3):497–50


