May 1, 2013
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
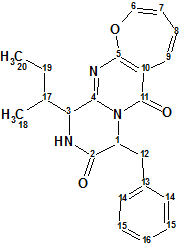
Protuboxepin A
Fungi have proven to be valuable resources for the discovery of novel secondary metabolites. Because the marine environment provides unique ecosystems and living conditions, marine fungi have been recognized as a potential source of diverse novel secondary metabolites. Lee et al [1] have investigated the chemical constituents of the extracts obtained from cultures of the marine-derived fungus Aspergillus sp. SF-5044. This study led to the isolation of new natural products, particularly, an oxepin-containing diketopiperazine-type metabolite named Protuboxepins A (1).
1
Protuboxepin A (1) was assigned the molecular formula C22H23N3O3 on the basis of HRESIMS data (m/z 378.1824 [M + H]+), which was fully supported by the 1H and 13C NMR data. Spectroscopic data (13C, 1H NMR, COSY, HMBC) used for the Protuboxepin A structure elucidation are collected in Table 1 (COSY data were not tabulated in the article [1], so they were determined from the spectral picture included in the Supporting Information). A slightly modified Molecular Connectivity Diagram (MCD) is presented in Figure 1.
Table 1. Protuboxepin A. Spectroscopic NMR data.
| Label | δC | δC calc | CHn | δH | M(J) | COSY | C HMBC |
| C 1 | 58.4 | 57.23 | CH | 5.34 | u | 3.35 | C 11, C 2, C 12, C 13, C 4 |
| C 2 | 169.3 | 169.43 | C | ||||
| C 3 | 59.4 | 51.42 | CH | 2.75 | u | 2.13 | C 4, C 2, C 5, C 17, C 19, C 18 |
| C 4 | 157.7 | 148.49 | C | ||||
| C 5 | 164.4 | 158.09 | C | ||||
| C 6 | 144.6 | 143.59 | CH | 6.08 | d(5.5) | C 5, C 7, C 8, C 9 | |
| C 7 | 118.4 | 120.12 | CH | 5.72 | u | C 6, C 9 | |
| C 8 | 129.3 | 124.55 | CH | 6.21 | u | 6.73 | C 10, C 6, C 7 |
| C 9 | 126.1 | 130.77 | CH | 6.73 | d(11.4) | 6.21 | C 11, C 7, C 6, C 5, C 10 |
| C 10 | 111.4 | 110.63 | C | ||||
| C 11 | 162.7 | 160.7 | C | ||||
| C 12 | 37.8 | 36.86 | CH2 | 3.35 | u | 5.34 | C 14, C 13, C 2, C 1 |
| C 13 | 135.8 | 134.77 | C | ||||
| C 14 | 130.9 | 128.71 | CH | 6.93 | d(7.0) | C 12, C 15 | |
| C 15 | 129.9 | 128.9 | CH | 7.25 | t(7.7) | C 13, C 16 | |
| C 16 | 129.1 | 128.48 | CH | 7.32 | t(7.3) | C 14 | |
| C 17 | 38.8 | 39.32 | CH | 2.13 | u | 1.14, 2.75, 0.84 | C 3, C 20, C 18, C 19, C 4 |
| C 18 | 15.4 | 14.19 | CH3 | 0.84 | d(7.0) | 2.13 | C 17, C 19, C 3 |
| C 19 | 24.8 | 25.36 | CH2 | 1.14 | u | 0.75, 2.13 | C 3, C 17, C 18, C 20 |
| C 20 | 12.3 | 11.22 | CH3 | 0.75 | t(7.3) | 1.14 | C 19, C 17 |
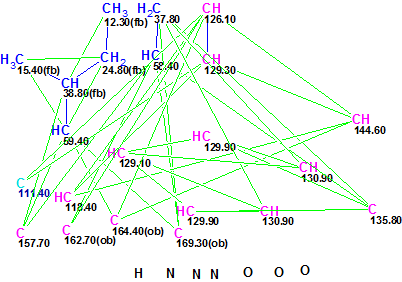
Figure 1. Protuboxepin A. Slightly modified Molecular Connectivity Diagram (MCD).
MCD overview. Only three atom property refinements were made at the MCD: a label “ob” (a sign of the presence of at least one hetereoatom in the first sphere of the given atom environment) was assigned to carbon atoms C 162.70, C 164.40 and C 169.30. Properties of the light blue carbon atom C 111.4 were left without any edits: along with C(sp2), the presence of O-C-O and N-C-O fragments is also conceivable. The numbers of hydrogen atoms attached to the carbons neighboring with a given CHn atom were set in accordance to 1H multiplicities shown in Table 1 (column M/J). The assigned hydrogen numbers can be seen when the cursor is placed on the corresponding carbon atom. The molecule contains one exchangeable proton, and if the IR spectrum of the sample diluted in CCl4 was available, the choice between OH and NH functionalities would be possible. Unfortunately no IR spectroscopic data were presented in the article [1], therefore an attempt to resolve the alternative was made using the so called methodology of “generalized portrait” (see [2]).
For this goal, a Fragment Search in the ACD Fragment Library by the 13C NMR spectrum of the compound was carried out, which resulted in selection of 1029 fragments. The following command was then activated: Structure Elucidation/Advanced/Search Functional Groups. This ranked the numbers of fragments containing given functional groups in decreasing order. Figure 2 shows twenty top structures and a row (#41–#45), the latter containing functional groups whose presence in the molecule was rejected by the program.
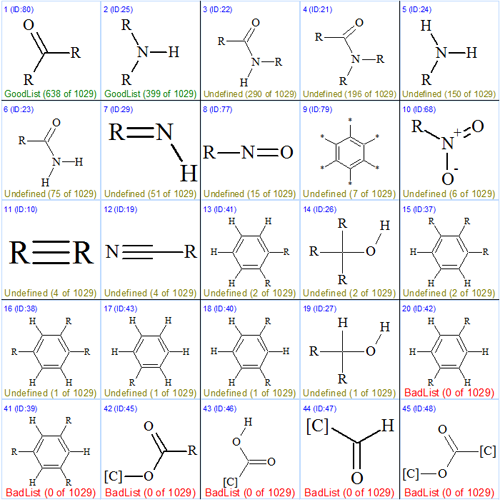
Figure 2. Protuboxepin A. Top of the Functional Group Library where functional groups are ranked in the order of decreasing numbers of fragments containing given substructures. The last row shows functional groups assigned by the program to “BadList”.
The first two functional groups are ones that are the most frequently presented in Found Fragments, and are recommended by the program to be included into User GoodList. Because both carbonyl and secondary amine groups are embedded into the secondary amid group (item #3) the fragment was placed into the User GoodList Library of Structure Elucidator. Note that only 2 of 1029 fragments contain secondary and tertiary alcohol functionality (items #14 and #19), which can be considered as an additional confirmation of the hypothesis about NH (not OH!) presence in the molecule. Moreover, the last row of the displayed Functional Groups can serve as a reason to exclude the presence of ester groups in the molecule.
MCD checking indicated that the 2D NMR data contain at least one nonstandard connectivity, so Fuzzy Structure Generation was initiated with the following parameters: m=1-20, a=1, “Stop Generation when Structures Generated”, calculate 13C chemical shifts during the structure generation. The results were: k=1928→1, tg = 5 m 40 s, 2 out of 35 connectivities were extended during generation, 160 from 595 possible connectivity combinations were used, dA=2.37, dN=1.82, dI=3.5.
Only a single structure (1a) was generated, which is identical to Protuboxepin A (1). The generated structure 1a is presented below along with 13C chemical shift assignments and nonstandard connectivities (red arrows).
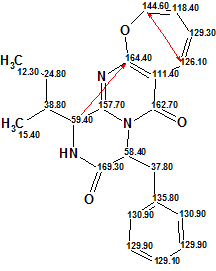
1a
This example demonstrates how using the “generalized portrait” methodology, i.e., a simple and straightforward elucidation process, quickly generated a single and correct solution to the problem.
References
- S. U. Lee, Y. Asami, D. Lee, J.-H. Jang, J. S. Ahn, H. Oh. Protuboxepins A and B and Protubonines A and B from the Marine-Derived Fungus Aspergillus sp. SF-504 J. Nat. Prod., 74(5):1284–1287, 2011.
- M.E. Elyashberg, A.J. Williams, K.A. Blinov. Contemporary Computer-Assisted Approaches to Molecular Structure Elucidation, Cambridge, RSC Publishing, p.482, 2012


