February 1, 2023
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
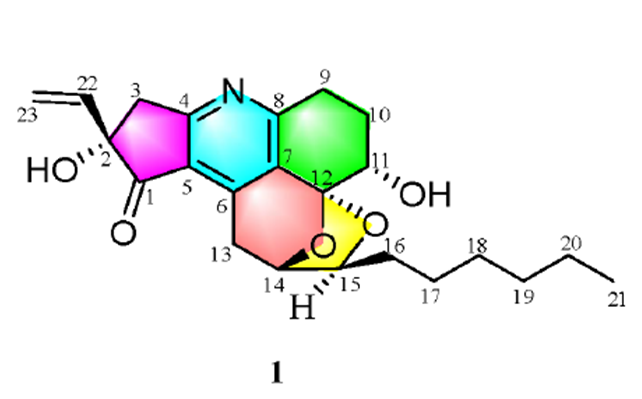
Trichopsistide A
Amongst the high variety of species found in soils, plants and decaying vegetation are Trichoderma. They can be found worldwide in various habitats. Many natural products have been isolated from Trichoderma, including polycyclic trichodermatides, which are natural polyketides with unusual skeletons. In total, three polycyclic trichodermatides have been identified and only trichodermatide A has a very unique pentacyclic skeleton containing a ketal, with a 6/6/6/6/5 ring system. Trichoderma koningiopsis WZ-196 from Rubia podantha Diels was investigated [1] as part of continued studies and search for structurally unique secondary metabolites of fungi.
As a result, a highly oxygenated ketal-containing pentacyclic polyketide, trichopsistide A (1) was isolated and identified. Its structure was determined by comprehensive spectroscopic analysis, CASE software and DFT chemical shift calculations. Compound 1 is the first example of a 5/6/6/6/5 pentacyclic ketal-containing polyketide pyridine alkaloid [1].
Trichopsistide A was obtained as a yellow solid. The molecular formula was found to be C23H29NO5 with 10 degrees of unsaturation on the basis of its 13C NMR and HRESIMS data (m/z 400.21165 [M + H]+; calcd 400.21185). The IR spectrum showed absorption bands at 3400, 2953, 2917, 2850, 1718, 1671, 1595, 1509, 1318, and 932 cm−1. Using the traditional method of structure elucidation based on analysis of HSQC, HMBC and COSY data the authors [1] deduced a large part of the molecular structure, but the lack of carbons bearing hydrogen atoms prevented them to assemble the full structure. To alleviate this problem, ACD/Structure Elucidator was utilized in [1]. The raw spectroscopic data were entered into the software and peak picking in 1D and 2D NMR spectra was undertaken. The data resulting from this are presented in Table 1.
Table 1. NMR spectroscopic data
| C/X Label | δC | δCcalc (HOSE) | XHn | δH | COSY | H to C HMBC |
| C 1 | 204.9 | 208.72 | C | |||
| C 2 | 81.2 | 81.96 | C | |||
| C 3 | 45.1 | 42.96 | CH2 | 3.22 | ||
| C 3 | 45.1 | 42.96 | CH2 | 3.42 | C 5, C 4, C 1 | |
| C 4 | 170.7 | 175.39 | C | |||
| C 5 | 125.7 | 118.58 | C | |||
| C 6 | 145.8 | 146.12 | C | |||
| C 7 | 131 | 129.16 | C | |||
| C 8 | 163.3 | 164.08 | C | |||
| C 9 | 28.2 | 27.04 | CH2 | 3.19 | 2.22 | C 11, C 7, C 8 |
| C 9 | 28.2 | 27.04 | CH2 | 2.93 | ||
| C 10 | 26.8 | 24.66 | CH2 | 2.28 | ||
| C 10 | 26.8 | 24.66 | CH2 | 2.22 | 3.19, 4.10 | |
| C 11 | 69.3 | 73.33 | CH | 4.1 | 2.22 | C 12, C 7 |
| C 12 | 104.5 | 111.15 | C | |||
| C 13 | 33.5 | 28.73 | CH2 | 3.54 | 4.6 | C 5, C 7, C 6, C 1 |
| C 13 | 33.5 | 28.73 | CH2 | 3.17 | ||
| C 14 | 78.2 | 77.64 | CH | 4.6 | 3.54, 3.98 | C 12, C 6 |
| C 15 | 83.1 | 80.53 | CH | 3.98 | 1.60, 4.60 | C 17, C 12 |
| C 16 | 36.1 | 31.95 | CH2 | 1.6 | 1.37, 3.98 | C 18 |
| C 17 | 26.5 | 25.29 | CH2 | 1.37 | 1.34, 1.60 | |
| C 18 | 30.5 | 28.75 | CH2 | 1.34 | 1.32, 1.37 | |
| C 19 | 33.1 | 31.4 | CH2 | 1.32 | 1.34 | C 17 |
| C 20 | 23.8 | 22.1 | CH2 | 1.33 | 0.92 | |
| C 21 | 14.6 | 14.1 | CH3 | 0.92 | 1.33 | C 20, C 19 |
| C 22 | 139.3 | 138.16 | CH | 5.97 | 5.25 | C 3, C 2, C 1 |
| C 23 | 116 | 115.49 | CH2 | 5.43 | ||
| C 23 | 116 | 115.49 | CH2 | 5.25 | 5.97 | C 2 |
The program created a Molecular Connectivity Diagram (MCD). And a carbonyl bond was manually added for the carbon at 204.9 ppm (Figure 1). Despite the absence of triple bond indicative bands in the IR spectrum, the authors allowed the presence of triple bonds in the generated structure (the corresponding option was marked in the dialog window “Create MCDs Options“.
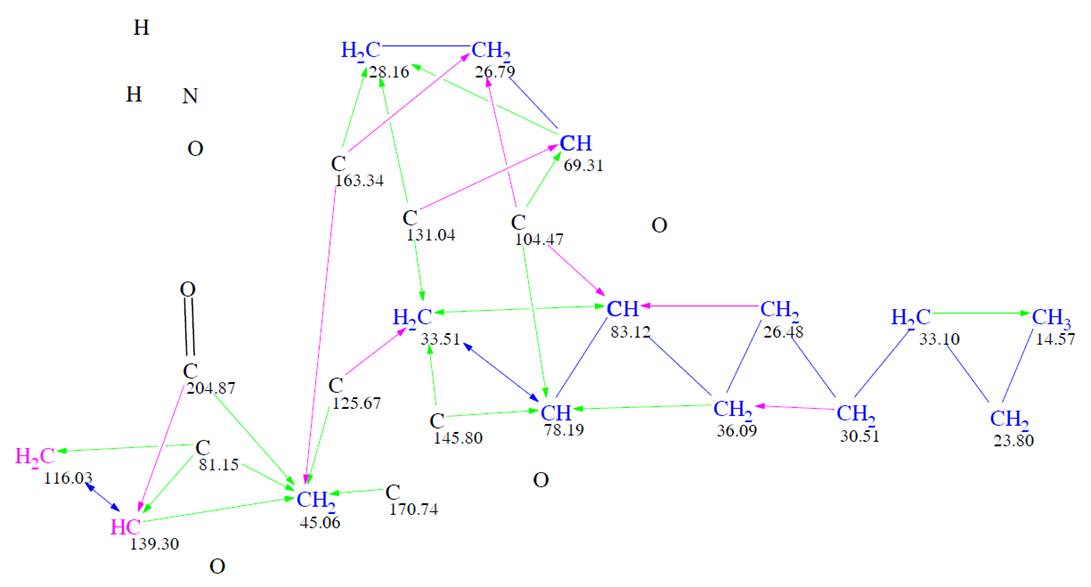
Figure 1. Molecular Connectivity Diagram. Hybridizations of carbon atoms are marked by corresponding colors: sp2 – violet, sp3 – blue, not defined – black. Green arrows mark HMBC correlations, blue arrows – COSY, violet arrows – 2D NMR correlations of nonstandard (longer) length.
Black atoms appeared in MCD due to the assumption that triple bonds are possible in the molecule, while nonstandard, longer, correlations (NSCs) were set by the program by taking into account the peak intensities in HMBC and COSY spectra. As a result of strict structure generation, 613,609 isomers were generated in 27 min, and 249 candidates structures were saved after spectral filtering. Then 13C chemical shifts were predicted for the structures generated using three algorithms implemented into ACD/SE – incremental, neural networks and HOSE code-based, and the output file was ranked in increasing order of average deviations. The top ten ranked structures found in [1] are shown in Figure 2.
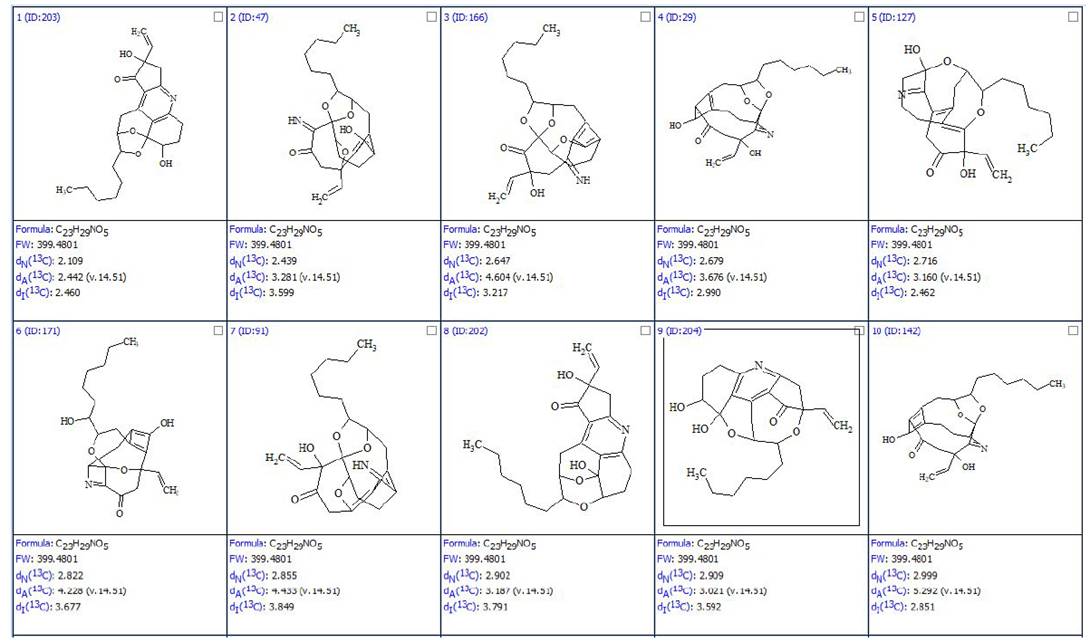
Figure 2. The top ten ranked structures of the output file. The average deviations of 13C chemical shifts determined by HOSE code, neural networks and incremental approach are denoted as dA, dN and dI correspondingly. Atom numbering is not shown.
The first ranked structure is the most probable one and it was assigned to trichopsistide A. DFT based 13C chemical shift prediction confirmed the assignment.
It was interesting to repeat the structure elucidation using the same data presented in Table 1 but with two tweaks:
- The intensities of HMBC and COSY peaks were set as unknown and all correlations were considered of standard length (2-3JHH,CH)
- The maximum bond multiplicity was set as 2
The corresponding MCD is shown in Figure 3.
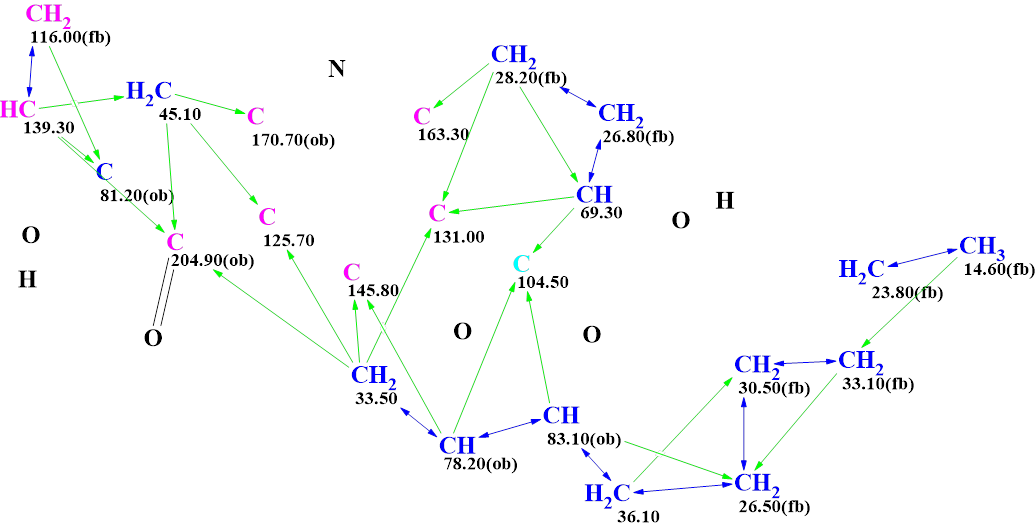
Figure 3. The MCD used for the recalculation. The atoms are placed in accordance with their positions in structure 1.
Checking the MCD for 2D NMR data consistency was finished with the following program message: “The minimum number of non-standard connectivities is 1”. That is, the program informed that at least one nonstandard correlation was detected in HMBC and COSY spectra. Therefore, Fuzzy Structure Generation (FSG) accompanied by 13C chemical shift prediction and structure filtering was initiated, while the options of FSG were determined automatically. Results: k = 37,221 → (structure filtering) → 559 → (duplicate removal) → 558, tg = 1 m 7 s.
The top four ranked structures are shown in Figure 4.
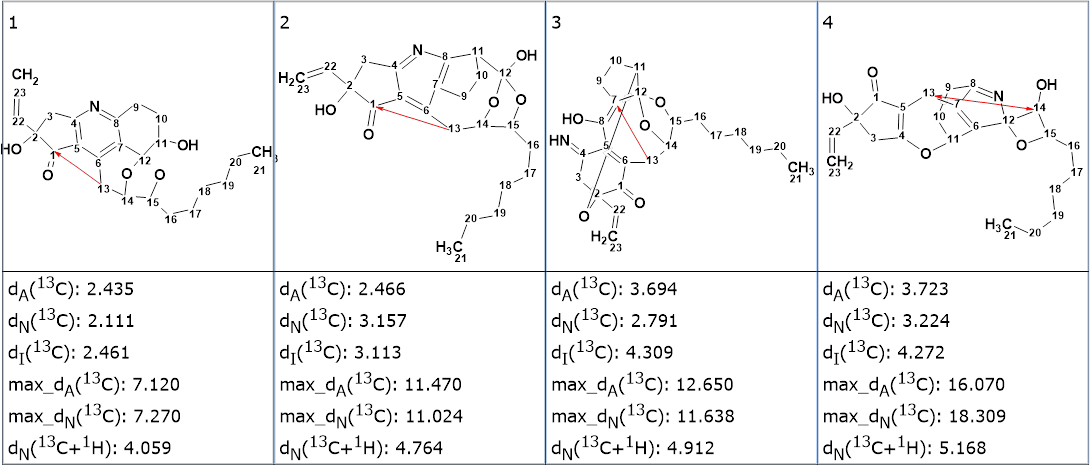
Figure 4. The top four ranked structures of the output file generated from the MCD, in Figure 3. The red arrow shows a correlation of nonstandard length.
We see that the best structure is the same as in Figure 2, and the program identified one NSC during FSG and lengthened it to allow for structure generation. The example shows that admitting the possibility of a triple bond presence in the molecule and automatic assignment of correlation lengths in HMBC and COSY spectra can lead to a noticeable increase of the generation time and even several closely matched resulting structures. We believe that utilization of the IR spectrum is very useful to understand whether a triple bond is present or not in the structure under investigation.
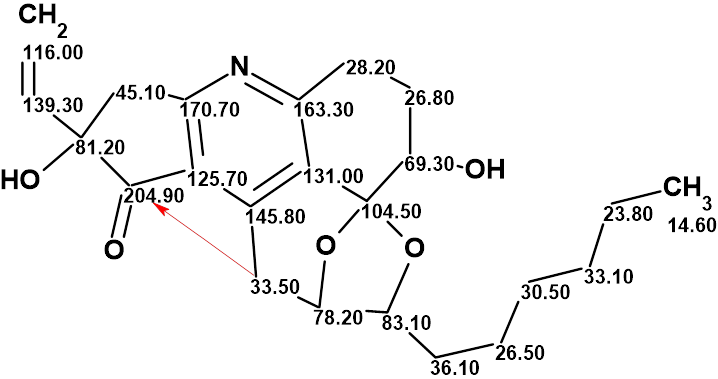
The structure of trichopsistide A with the assigned 13C chemical shifts is shown below:
References
- L. Feng, A.-X. Zhang, R.-R Shang, X.-J. Wang, N.-H. Tan, Z. Wang. (2022). Trichopsistides A and B: Two Highly Oxygenated Pentacyclic Polyketides with Promising Inhibitory Effects on the NF-κB Signaling Pathway from the Fungus Trichoderma koningiopsis WZ-196. J. Org. Chem., 87, 14058−14067.