April 1, 2012
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Psychotripine
The Initial Challenge
A recent article1 highlighted the isolation and structure elucidation of a new natural product Psychotripine, which exhibited a complex carbon skeleton comprised of 11 rings. The 2D NMR data published for this molecule were limited. In addition the molecule contains a quaternary carbon with no neighboring heteroatoms and an anomalous 13C chemical shift at 69.1 ppm. The purpose of this challenge was to attempt the Computer-Assisted Structure Elucidation (CASE) of Psychotriptine using only the original limited and incomplete NMR data gathered from the original publication.

Figure 1
The Evidence
- The molecular formula of Psychotripine was obtained as C33H34N6 with 20 degrees of unsaturation. The C:H ratio indicates a deficit of hydrogen.
- In the original publication, only the 13C and 1H NMR spectra were presented in a table, while the 2D NMR data were not tabulated. Only key HMBC (green arrows) and COSY (violet bonds) correlations were shown on the structure as shown in Figure 2.
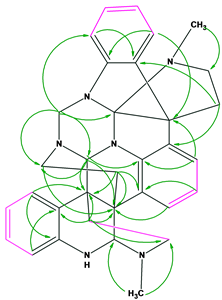
Figure 2
The CASE (Computer Assisted Structure Elucidation) Investigation
To solve this problem using ACD/Structure Elucidator, the 13C chemical shifts and the key correlations highlighted in Figure 2 were entered into the CASE program. It was suggested that the quaternary carbon C at 106.9 ppm existed in the sp3 hybridization and was connected with 3 nitrogen atoms, because the molecule contains an even number of sp2 carbons (18 aromatic atoms). The suggestion was confirmed by 13C chemical shift prediction for several model molecules containing sp3 hybridized carbon in the same environment.
Based on this information, the molecular connectivity diagram (MCD) was created for structure generation in ACD/Structure Elucidator (Figure 3).
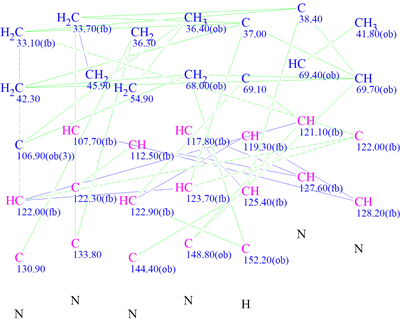
Figure 3: The Molecular Connectivity Diagram produced by Structure Elucidator
With limited and incomplete 2D NMR information, a large number of candidate structures was generated. As a result a filter was introduced in the process to reject any structure candidates with calculated 13C chemical shifts that deviated with more than 4 ppm from the experimental data (that is d>4 ppm, where d is average deviation between experimental and calculated spectra). After 90.5 hours, 16 unique chemical structures were generated (3.7 million without filter).
After ranking the candidates by 13C NMR predictions, the structure of Psychotripine as suggested by the authors1 was ranked in the first position. The top 3 structures with their deviations are presented in Figure 4.
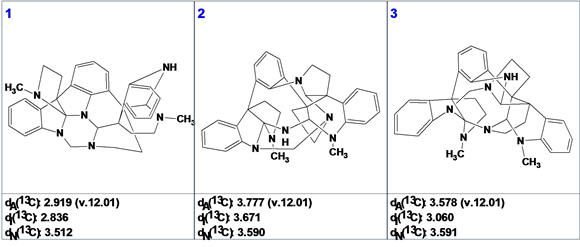
Figure 4: The top 3 structures of the ranked output file. Designations of deviations: dA—HOSE code based chemical shift calculation, dI—calculation based on incremental approach, dN—calculation based on artificial neural nets algorithm.
Therefore, the structure generation from limited 2D NMR data was automatically performed during 90.5 h without any user time consumption.
Conclusions
ACD/Structure Elucidator generated the correct structure and selected it among ~3.7 million generated structures. With the usage of a chemical shift prediction filter only 16 structures were highlighted, and the correct structure was revealed after 90 hours without any supervision or intervention.
Note that the large processing time is the payment for obtaining the correct structure based on limited and incomplete NMR data. This study shows that even with missing information, the software is powerful enough to elucidate the correct chemical structure of very complex molecules.
ACD/Structure Elucidator is a complete elucidation package offering tools to speed up the elucidation process and ensure that no candidate is overlooked.
References
- X.-N. Li, Y. Zhang, X.-H. Cai et al. Psychotripine: A New Trimeric Pyrroloindoline Derivative from Psychotria pilifera. Org. Lett., 13:5896-5899, 2011.


