February 1, 2016
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Ruthmycin
Pyranonaphthoquinones are polyketides produced by various bacteria and fungi, the majority of which have been isolated from actinomycetes. The frenolicins are a predominate group of pyranonaphthoquinones known for their antibacterial, antifungal, and antiprotozoal activities.
Wang et al [1] have detected a new metabolite that displayed a distinct LC-MS signature from known frenolicins of Streptomyces sp. Subsequent isolation, purification, and structure elucidation of the newly identified metabolite revealed a new tetracyclic skeletal framework, henceforth referred to as Ruthmycin (structure 1) in reference to the producing strain’s point of origin.
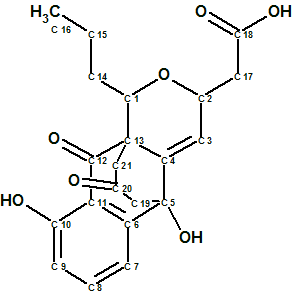
1
Ruthmycin possesses an unprecedented signature C3 bridge and a corresponding fused six-member ring. In the context of known frenolicin-type scaffolds, the presence of a C-3/C-4 double bond is also unprecedented.
Compound 1 was obtained as a yellow amorphous powder. HRESIMS data established the molecular formula of Ruthmycin as C21H22O7, indicating 11 degrees of unsaturation. The IR spectrum of compound 1 displayed characteristic absorptions for OH (3374 cm−1) and C=O (1717 cm−1) functionality. The structure of the Ruthmycin molecule was elucidated from 1H, 13C, HSQC, HMBC and COSY spectra. The corresponding data are collected in Table 1.
Table 1. Spectroscopic NMR data for Ruthmycin.
| X Label | δX | δC calc | XHn | δH | COSY | C HMBC |
| C 1 | 76.3 | 74.28 | CH | 4.09 | 1.55 | |
| C 2 | 66.5 | 71.95 | CH | 4.68 | C 3, C 4, C 17, C 18 | |
| C 3 | 121.8 | 125.08 | CH | 6.33 | C 2, C 4, C 5, C 13, C 17 | |
| C 4 | 140.3 | 142.78 | C | |||
| C 5 | 73 | 71.31 | C | |||
| C 6 | 149.8 | 142.81 | C | |||
| C 7 | 115.9 | 113.93 | CH | 7.35 | 7.65 | C 5, C 11 |
| C 8 | 139.3 | 136.92 | CH | 7.65 | 6.88, 7.35 |
C 6, C 10 |
| C 9 | 117.6 | 117.6 | CH | 6.88 | 7.65 | C 7, C 11 |
| C 10 | 163 | 162.94 | C | |||
| C 11 | 115.1 | 114.89 | C | |||
| C 12 | 205.3 | 203.78 | C | |||
| C 13 | 52.8 | 54.1 | C | |||
| C 14 | 29.2 | 34.95 | CH2 | 1.75 | ||
| C 14 | 29.2 | 34.95 | CH2 | 1.55 | 1.42, 4.09 |
|
| C 15 | 20 | 19.29 | CH2 | 1.64 | ||
| C 15 | 20 | 19.29 | CH2 | 1.42 | 0.92, 1.55 |
C 16 |
| C 16 | 14.1 | 13.97 | CH3 | 0.92 | 1.42 | C 14, C 15 |
| C 17 | 39.9 | 40.22 | CH2 | 2.72 | C 2, C 3, C 18 |
|
| C 18 | 171.9 | 175.03 | C | |||
| C 19 | 60 | 51.33 | CH2 | 2.61 | C 4, C 5, C 6, C 20 | |
| C 19 | 60 | 51.33 | CH2 | 3.02 | ||
| C 20 | 204.7 | 205.52 | C | |||
| C 21 | 54.5 | 45.1 | CH2 | 2.58 | C 1, C 12, C 19 | |
| C 21 | 54.5 | 45.1 | CH2 | 3.33 | ||
| O 1 | 100* | OH | 6.46 | C 4, C 5, C 6, C 19 | ||
| O 2 | 150* | OH | 12.15 | C 10, C 11 |
* Fictitious 17O chemical shifts.
The data from Table 1 were input into the ACD/Structure Elucidator Suite software, which created a Molecular Connectivity Diagram (MCD, see Figure 1).
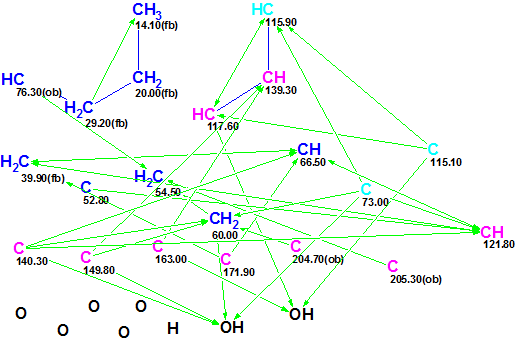
Figure 1. Molecular Connectivity Diagram (MCD) for Ruthmycin.
MCD overview. Figure 1 shows that the hybridization state of three carbon atoms (C 73.00, C 115.10 and C 115.90, marked by light blue color) was set as sp3 or sp2 (i.e., not sp) by the software, and are denoted by blue and violet colors in the MCD. The label fb (forbidden) means that a heteroatom cannot be a neighbor of a given carbon, while the label ob (obligatory) denotes that a neighboring heteroatom is required.
No user edits were made to the MCD. The software determined that there were no contradictions in 2D NMR data, and consequently Strict Structure Generation was initiated. This process was accompanied with 13C chemical shift prediction, and those structures which were characterized by an average deviation greater than 5 ppm were automatically rejected by the system. The following results were obtained: k = 4205 → 26 → 23, tg = 30 s. After 13C chemical shift prediction by three methods (Incremental, Neural Networks, HOSE code based) the output structures were ranked in ascending order of the average deviation dA (13C chemical shifts were calculate by HOSE code based approach). The six top structures of the ranked file are presented in Figure 2.
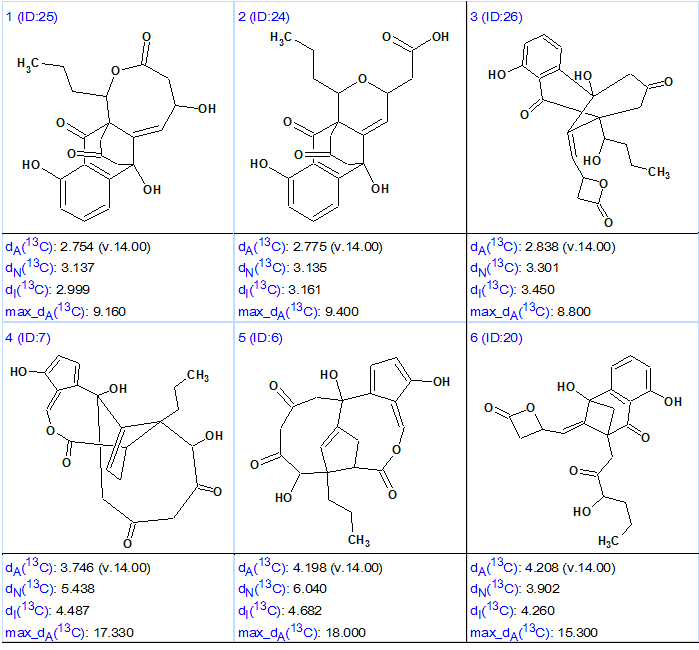
Figure 2. Six top structures of the ranked output file for Ruthmycin.
Figure shows that the first two structures (#1 and #2) are characterized by very similar average and maximum deviations, while structure #2 coincides with the original structure 1 suggested for Ruthmycin. The main difference between structures #1 and #2 is in the presence of a COOH group in structure #2, is absent from structure #1. This acid group is known to have a very specific characteristic feature in IR spectrum—a monomer absorption band at 3500–3550 cm-1 in a diluted neutral solvent. Therefore, if this additional experiment was carried out the choice between structures #1 and #2 would be done confidently. Structure #3 can be deemed too exotic for a natural product, and the deviations calculated for the remaining structures are very large, discounting them as candidates.
Though the “resolving power” of 1H chemical shift prediction as an aid for selecting the preferable structure is rather modest, in many cases it can help to confirm (but not prove) some structural hypotheses. Figure 3 shows structures #1 and #2 with 1H chemical shift assignments and average deviations dA(1H) calculated by the HOSE code-based approach. Colored areas around the atoms show accuracy of 1H chemical shift calculation.
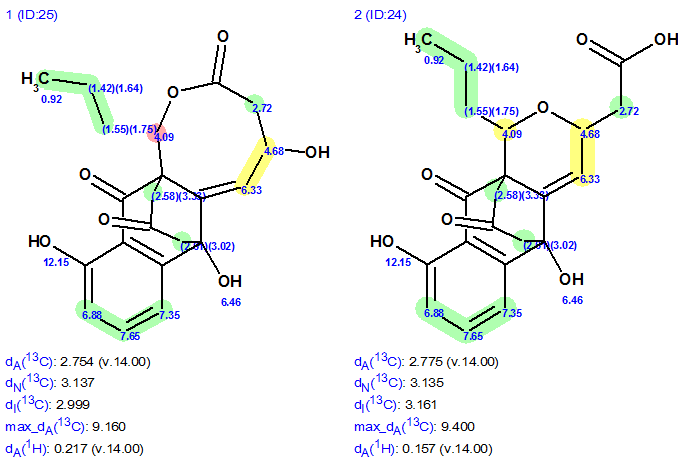
Figure 3. Structures #1 and #2 with 1H chemical shift assignments and average deviations dA(1H) calculated by HOSE code based approach.
Figure 3 shows that the structure #2 is more preferable because the average predicted 1H chemical shift deviations of #2 are lower (dA#2, 1H < dA#1,1H) and the difference between experimental chemical shift 4.09 ppm (red cycle) and predicted one is too large (1.17 ppm in this case).
We see that the elucidation of the unprecedented structure of Ruthmycin was performed in fully automatic mode, but there is a simple additional experiment—to obtain an IR spectrum from diluted sample—needed for a final structure endorsement. The structure of Ruthmycin supplied with 13C chemical shift assignments is shown below:
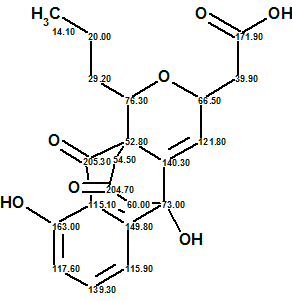
2
References
- X.Wang, S.I. Elshahawi, K. A. Shaaban, L. Fang, L.V. Ponomareva,Y. Zhang, G. C. Copley, J.C. Hower, C.-G. Zhan, M.K. Kharel, J.S. Thorson. (2014). Ruthmycin, a New Tetracyclic Polyketide from Streptomyces sp. Org. Lett., 16: 456–459.


