February 1, 2014
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Schizocommunin
Schizocommunin was first reported in 1999 from the liquid culture medium of Schizophyllum commune, strain IFM 46788 (monokaryon). The following structure was suggested for schizocommunin:
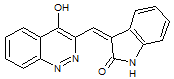
1
Biological studies on schizocommunin were prevented by the very limited supply of schizocommunin available from natural sources, and there have been no reports of the total synthesis of schizocommunin. Although a spectroscopic analysis revealed that the structure of schizocommunin included both 4-hydroxycinnnoline and oxindole skeletons that were connected by exomethylene, as shown in (Z)-1hydroxy in Figure 1, the configuration of the C(3)-C(9′) olefin was not discussed in detail. It is possible that 1 has either geometric structures (Z)- and (E)-1-hydroxy or tautomeric structures (Z)- and (E)-1-keto (Figure 1).
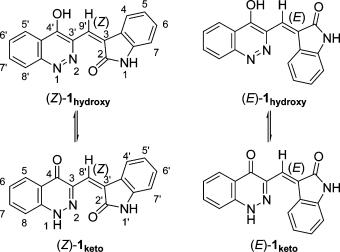
Figure 1. Configurations of schizocommunin.
Recently Uehata and co-workers [1] performed the total synthesis of the putative structures of schizocommunin (Z)-1hydroxy and its geometric isomer (E)-1hydroxy, which both exist in a keto form. However, the 1H NMR spectra for synthetic (Z)- and (E)-1keto were not identical to those reported for natural schizocommunin. A reinvestigation of the NMR and IR results for natural schizocommunin led the researchers to propose a revised structure, quinazolinone 2, which was synthesized in a single step. All of the spectral data of (Z)-2 were identical to those reported for natural schizocommunin (Figure 2).
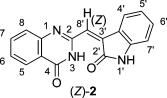
Figure 2
The authors [1] performed a careful investigation of the problem, as a result of which methods of synthesis of corresponding compounds were developed and the originally determined structure of schizocommunin was revised.
We suggested that if a CASE approach were used elucidate the structure when this compound was firstly isolated, the structure revision would be not necessary: for final structure confirmation X-ray crystallographic analysis would be sufficient.
With this in mind, the 1D and 2D NMR data of schizocommunin was extracted from the Supporting Information of [1] and were entered into ACD/Structure Elucidator Suite. The data are shown in Table 1 where atom numbers correspond to those which are displayed in the structure below:
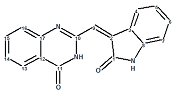
Table 1. Schizocommunin NMR spectroscopic data.
| Label | δC | δC calc | XHn | δH | M(J) | COSY | C HMBC |
| C 1 | 168.8 | 167.84 | C | ||||
| C 2 | 134.3 | 144.21 | C | ||||
| C 3 | 123.3 | 123.34 | C | ||||
| C 4 | 121.9 | 122.24 | CH | 7.94 | u | 7.1 | C 2, C 6, C 8 |
| C 5 | 122.6 | 121.41 | CH | 7.1 | u | 7.94, 7.36 | C 3 |
| C 6 | 131.6 | 129.37 | CH | 7.36 | u | 7.10, 6.93 | C 8, C 4 |
| C 7 | 110.6 | 110.36 | CH | 6.93 | u | 7.36 | C 8 |
| C 8 | 141.6 | 141.29 | C | ||||
| C 9 | 130 | 121.59 | CH | 7.58 | u | C 1, C 3, C 2 | |
| C 10 | 150.4 | 154.38 | C | ||||
| C 11 | 160.8 | 162.8 | C | ||||
| C 12 | 121.5 | 120.16 | C | ||||
| C 13 | 126 | 125.92 | CH | 8.17 | u | 7.6 | C 11, C 15, C 17 |
| C 14 | 128 | 128.3 | CH | 7.6 | u | 7.88, 8.17 | C 12 |
| C 15 | 134.7 | 135.01 | CH | 7.88 | u | 7.60, 7.79 | C 13, C 17 |
| C 16 | 128 | 124.54 | CH | 7.79 | u | 7.88 | C 12, C 14 |
| C 17 | 148.9 | 148.45 | C | ||||
| N 1* | 150 | NH | 11.49 | u | |||
| N 2* | 170 | NH | 13 | u |
*Fictitious 15N chemical shifts.
A Molecular Connectivity Diagram was created (MCD, see below), where two isolated groups of carbon atoms were selected, allowing the recognition of two benzene rings. The carbons included in benzene rings were connected by COSY-like connectivities. As the presence of two carbonyl absorption bands at 1675 and 1685 cm-1 are evident from the IR spectrum, carbons C 160.8 and 168.8 were connected to oxygens with double bonds.
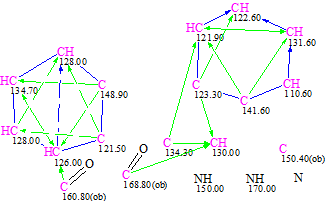
Figure 3. MCD of schizocommunin.
Structure generation was performed with the option “Allow Bonds between Heteroatoms of the Same Atom Type” selected. 13C NMR chemical shifts were calculated during the structure generation, and the filtering procedure was used (structures characterized with d>5 ppm were rejected by the filter). The results were: k= 3614 → 44 → 22, tg = 2 s. Eight top structures of the output file ranked by dA deviations (HOSE code based spectrum prediction) are shown in Table 2.
Table 2. Eight top structures of the ranked output file of the first run.
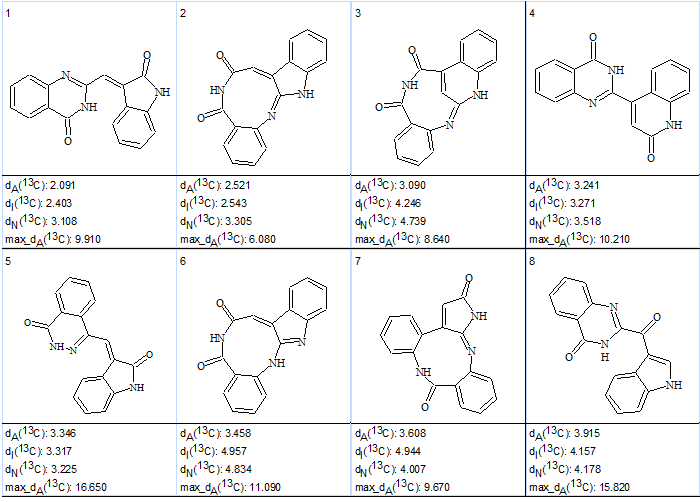
The table shows that the true structure of schizocommunin was selected as the most probable one by all three methods of chemical shift prediction. Z and E configurations of the molecule supplied with the 13C chemical shift assignment are displayed below:
Figure 4. Z and E configurations of schizocommunin.
It was also interesting to see how both structures—original and revised—will be ranked if both of them are generated from the MCD if N=N bonds are allowed. The correspondent MCD is shown below:
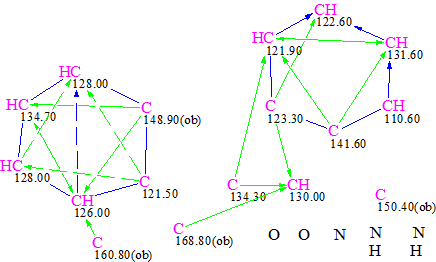
Figure 5. MCD of schizocommunin when N=N bonds are allowed
The structure generation under conditions similar to those used for the first run was carried out from this MCD, and the following results were obtained: k=176535 → 344 → 297, tg = 2 m 10 s. Eight top structures of the output file ranked by dA deviations (HOSE code based spectrum prediction) are shown in Table 3.
Table 3. Eight top structures of the ranked output file from the second run.
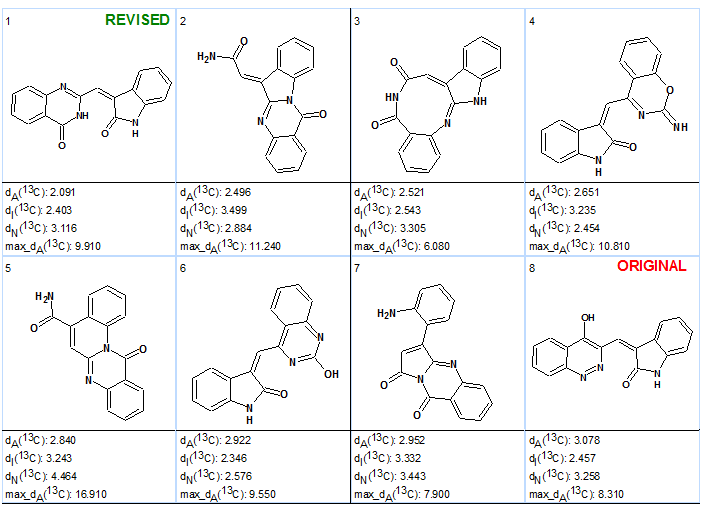
Table 3 shows that the original structure was placed on the 8th position by the ranking procedure, which evidently confirmed priority of the revised structure. Taking into account that all structures are very similar and the difference between deviations is less than 1 ppm, confirmation of the revised structure by X-ray analysis is desirable. This was done by the authors of article [1].
The example shows that application of Structure Elucidator system is capable of significant influencing strategy of structure revision based on chemical synthesis.
References
1. K. Uehata, N. Kimura, K. Hasegawa, S. Arai, M. Nishida, T. Hosoe, K. Kawai, A. Nishida. Total Synthesis of Schizocommunin and Revision of its Structure. J. Nat. Prod., 76:2034–2039, 2013.


