April 1, 2015
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Sinensilactam A
While searching for biologically active natural products from lingzhi mushrooms, Luo and coworkers [1] investigated G. Sinensis, a fungal species distributed in the eastern and southern regions of China. Many earlier studies of this traditional medicine revealed that it contains polysaccharides, triterpenoids, alkaloids, fatty acids, nucleotides, proteins, peptides, trace elements, sterols, and ganosinensins A—C, the latter of which are hybrids of a triterpenoid and a prenylated phenols. Despite these observations, the presence of meroterpenoids in this species remained largely unknown. The authors’ efforts focusing on lingzhi meroterpenoids from G. sinensis led to the isolation of a meroterpenoidal hybrid metabolite, called Sinensilactam A (see Figure 1 and structure 1), which contains a rare 2H-pyrrolo[2,1-b][1,3]- oxazin-6(7H)-ring system. Structure 1 was confirmed by X-ray crystallographic analysis.
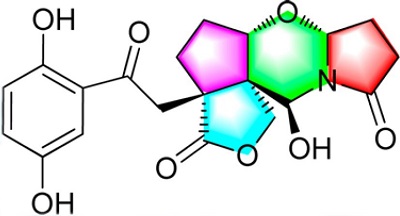
Figure 1. Sinensilactam A.
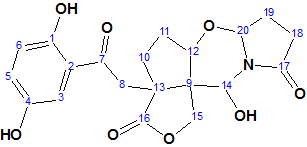
1
Sinensilactam A (l), obtained from G. sinensis as colorless crystals (MeOH), has the molecular formula C20H21NO8 (11 degrees of unsaturation), based on analysis of its HRESIMS, 13C NMR, and DEPT spectra. The spectroscopic data used for the molecular structure elucidation with the aid of ACD/Structure Elucidator Suite are presented in Table 1.
Table 1: Spectroscopic NMR data in DMSO-d6 for Sinensilactam A.
| Label | δC | δC calc | XHn | δH | Mult. | COSY | C HMBC |
| C 1 | 153.2 | 157.12 | C | ||||
| C 2 | 120 | 121.04 | C | ||||
| C 3 | 114.6 | 115.64 | CH | 7.22 | s | C 5, C 1 | |
| C 4 | 149.5 | 150.22 | C | ||||
| C 5 | 124.3 | 125.01 | CH | 6.99 | d | 6.83 | C 1 |
| C 6 | 118.5 | 119.17 | CH | 6.83 | d | 6.99 | C 7, C 4 |
| C 7 | 201.6 | 197.71 | C | ||||
| C 8 | 45.4 | 43.99 | CH2 | 3.41 | u | C 16, C 13, C 10, C 7, C 2 | |
| C 8 | 45.4 | 43.99 | CH2 | 3.85 | u | ||
| C 9 | 51.3 | 54.87 | C | ||||
| C 10 | 33.9 | 32.07 | CH2 | 1.85 | u | ||
| C 10 | 33.9 | 32.07 | CH2 | 1.8 | u | 1.9 | C 16, C 13, C 9 |
| C 11 | 25 | 29.64 | CH2 | 1.55 | u | ||
| C 11 | 25 | 29.64 | CH2 | 1.9 | u | 4.32, 1.80 | C 9 |
| C 12 | 77.1 | 73.15 | CH | 4.32 | u | 1.9 | C 20, C 15, C 9 |
| C 13 | 48.2 | 54.36 | C | ||||
| C 14 | 75.9 | 75.71 | CH | 5.45 | u | 6.53 | C 15, C 12, C 20, C 9, C 17 |
| C 15 | 69.2 | 68.02 | CH2 | 3.76 | u | ||
| C 15 | 69.2 | 68.02 | CH2 | 4.24 | u | C 13, C 9, C 16 | |
| C 16 | 181.4 | 179.59 | C | ||||
| C 17 | 172.6 | 175.38 | C | ||||
| C 18 | 29.3 | 29.14 | CH2 | 2.29 | u | ||
| C 18 | 29.3 | 29.14 | CH2 | 2.39 | u | 1.73 | C 17 |
| C 19 | 23.5 | 27.78 | CH2 | 1.73 | u | 2.39, 5.25 | |
| C 19 | 23.5 | 27.78 | CH2 | 2.25 | u | ||
| C 20 | 84.5 | 84.81 | CH | 5.25 | u | 1.73 | C 17 |
| O 1 | OH | 10.7 | s | ||||
| O 2 | OH | 9.2 | s | ||||
| O 3 | OH | 6.53 | u | 5.45 |
A Molecular Connectivity Diagram (MCD), slightly edited in accordance with characteristic chemical shifts in 13C and 1H NMR spectra, is presented in Figure 2.
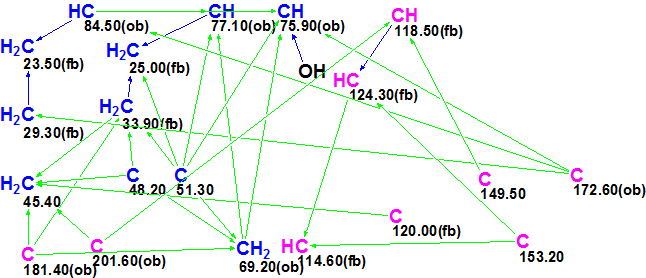
Figure 2. Slightly edited Molecular Connectivity Diagram for Sinensilactam A.
Diagram overview. Atom C 201.60 and carbons 69.20, 75.90, 77.10 and 84.50 were automatically supplied with the label “ob” (obligatory), which means that a heteroatom should exist in the first sphere of a carbon atom environment. Taking into account the number of heteroatoms (9), the same label was assigned to atoms C 172.60 and C 181.40 by the user. Label “fb” (neighbor heteroatom is forbidden) was set by the user for four sp2-hybridized carbons with 13C chemical shifts lying in the interval between 114.60 and 124.3 ppm. The number of hydrogens attached to the carbon atoms which are located in the first sphere of environment of definite carbons were set in accordance with 1H signal multiplicities shown in Table 1 (“Mult.” column).
Checking the Molecular Connectivity Diagram for the presence of contradictions was completed with the following message from the software: “Current Molecular Connectivity Diagram (MCD) passed all tests. No updates performed”. As no nonstandard connectivities (NSC) were detected in 2D NMR data, Strict Structure Generation [2,3] accompanied with 13C spectrum prediction was performed. The generated structures for which average 13C deviations > 5 ppm were rejected. The results were: k=44 → 0, tg = 0.7 s. It turned out that all generated structures were rejected in accordance with the criterion d > 5 ppm. This is a hint to suggestion that there are latent NSCs that were not detected by the program.
Therefore the Fuzzy Structure Generation [2,3] was run with the options m=1 , a=1 (supposed number of NSCs is equal to one, and possible augmentation of the connectivity lengths is of one bond).
The results were: k = 144 → 16 → 2, tg = 1 m 55 s. The output structures ranked in order of average deviations increase are presented in Figure 3.
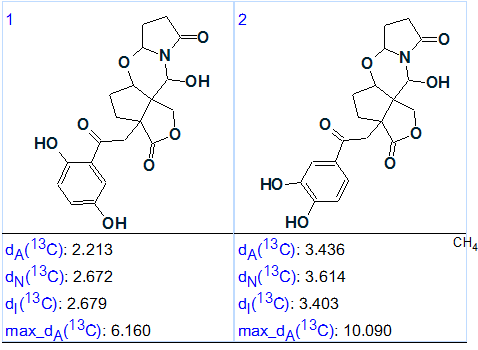
Figure 3. The ranked output file for Sinensilactam A.
We see that structure #1 in Figure 3 coincides with the structure 1 of Sinensilactam A as determined by authors. The structure with 13C chemical shift assignments produced by the program automatically is shown below (the red arrow shows the nonstandard connectivity detected in the HMBC data):
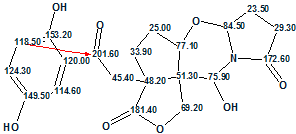
Therefore structure of Sinensilactam A containing a rare 2H-pyrrolo[2,1-b][1,3]- oxazin-6(7H)-ring system was reliably elucidated with the aid of ACD/Structure Elucidator Suite. In doing so, Fuzzy Structurer Generation allowed us to easily circumvent a latent contradiction existed in 2D NMR data.
References
- Qi Luo, Lei Tian, Lei Di, Yong-Ming Yan, Xiao-Yi Wei, Xin-Fang Wang, and Yong-Xian Cheng. (±)-Sinensilactam A, a Pair of Rare Hybrid Metabolites with Smad3 Phosphorylation Inhibition from Ganoderma sinensis. Org. Lett. 2015, ASAP, DOI: 10.1021/acs.orglett.5b00448, Publication Date (Web): March 6, 2015
- M.E. Elyashberg, A.J. Williams, K.A. Blinov, Contemporary computer-assisted approaches to molecular structure elucidation, RSC Publishing, Cambridge, 2012, 482 p.
- M.E. Elyashberg, A.J. Williams, Computer-based Structure Elucidation from Spectral Data. The Art of Solving Problems, Springer, Heidelberg, 2015, 446 p.


