October 1, 2015
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Spirochensilide A
The genus of Abies has been studied extensively because of structurally diverse and biologically active terpenoid constituents. More than 250 terpenoids and their analogs have been reported from this genus, and most of them are lanostane triterpenoids. Furthermore, several types of lanostane-related carbon skeletons have been found in the Abies genus.
A. chensiensis is one of the Chinese endemic plants mainly distributed over the Shanxi and Hubei provinces of the People’s Republic of China. As part of a program to search for unique and bioactive secondary metabolites, Zhao et al [1] isolated two novel rearranged triterpenoids from the leaves and twigs of A. chensiensis.—Spirochensilides A and B. The spectroscopic data used to elucidate the structure of Spirochensilide A (1), a compound containing unprecedented spiro-[5,6]system, were used to challenge ACD/Structure Elucidator Suite.
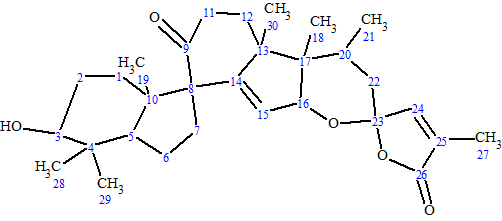
1
Spirochensilide A was obtained as a colorless crystal. The HRESIMS data suggested the molecular formula of 1 was C30H42O5 (m/z 483.3115 [M + H]+ calcd for 483.3105 [M + H]+), with 10 degrees of unsaturation. The IR spectrum exhibited absorptions at 3303 and 1736 cm-1, assignable to hydroxyl and carbonyl, respectively. The 13C NMR spectrum exhibited 30 carbon signals. To elucidate the structure, the authors [1] used 1D and 2D NMR data, including HSQC, HMBC, and COSY. The data are presented in Table 1.
| Label | δC | δC calc | XHn | δH | M(J) | COSY | C HMBC |
| C 1 | 149.2 | 156.25 | C | ||||
| C 2 | 159.5 | 158.5 | C | ||||
| C 3 | 124.5 | 122.54 | CH | 7.36 | d(8.5) | 7.87 | C 5 |
| C 4 | 135.2 | 135.39 | CH | 7.87 | d(8.5) | 7.36 | C 1, C 6, C 2 |
| C 5 | 130.4 | 128.85 | C | ||||
| C 6 | 92.7 | 87.76 | C | ||||
| C 7 | 73.2 | 65.87 | CH | 4.9 | s | C 5, C 13, C 8, C 6, C 9 | |
| C 8 | 138.6 | 136.39 | C | ||||
| C 9 | 126.8 | 127.99 | CH | 8.12 | d(1.0) | C 13, C 11, C 7, C 18 | |
| C 10 | 137.9 | 135.53 | C | ||||
| C 11 | 128.4 | 128.61 | CH | 8.01 | d(8.0) | 8.31 | C 9, C 13, C 18 |
| C 12 | 125.4 | 128.99 | CH | 8.31 | d(8.0) | 8.01 | C 10, C 8, C 1 |
| C 13 | 137.2 | 136.75 | C | ||||
| C 14 | 174.8 | 172.81 | C | ||||
| C 15 | 74.4 | 75.36 | CH | 4.61 | q(6.8) | 1.34 | C 14 |
| C 16 | 18.9 | 17.83 | CH3 | 1.34 | d(6.8) | 4.61 | C 14, C 15 |
| C 17 | 24.8 | 24.84 | CH3 | 2.57 | s | C 3, C 2 | |
| C 18 | 198.3 | 198.01 | C | ||||
| C 19 | 27.5 | 26.46 | CH3 | 2.62 | s | C 18 | |
| N 1 | 100* | NH | 9.1 | u | |||
| O 1 | 150* | OH | 6.27 | u |
*Fictitious 15N and 17O chemical shifts introduced to define dH for NH and OH groups.
The data presented in Table 1 were input into the program and a Molecular Connectivity Diagram (MCD) was created, to which the following minor edits were applied: two of four carbon atoms (66.9, 86.1, 105.4 and 132) for which hybridization not sp (sp3 or sp2) was assigned (indicated by light blue color) were labeled as C 66.9 – sp3 and C 132 – sp2/fb (“fb” means that connection to an heteroatom is forbidden); atom C 170.9 (sp2 hybridized) was marked as “ob” (neighboring of a heteroatom is obligatory), while atom C 214.9 was connected with an oxygen by double bond; finally, the numbers of hydrogen atoms attached to the carbons situated in the closest environment of CHn groups were set in accordance with signal multiplicities in the 1H NMR (see Table 1). Because atom CH 86.1 (4.51) may be in either sp3/ob or sp2/fb hybridization states, no edits of its properties were made; also no changes were also made regarding the properties of atom C 105.4 (either sp2/fb or sp3/ob for O-C-O). The slightly edited MCD is shown in Figure 1.
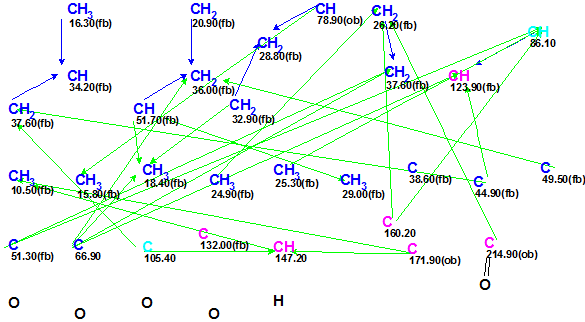
Figure 1. Slightly edited MCD for Spirochensilide A.
As no contradictions were detected in the 2D NMR data, strict structure generation followed by 13C chemical shift prediction and structure filtering was initiated. Structures characterized by deviations d(13C)>5 ppm were rejected during the structure generation. The following results were obtained: k = 1300 → 43 → 27, tg=11m 50s.
The six top structures of the output structural file, ranked by average 13C chemical shift deviations dA(13C), are shown in Figure 2. These deviations correspond to the HOSE code-based method of chemical shift prediction.
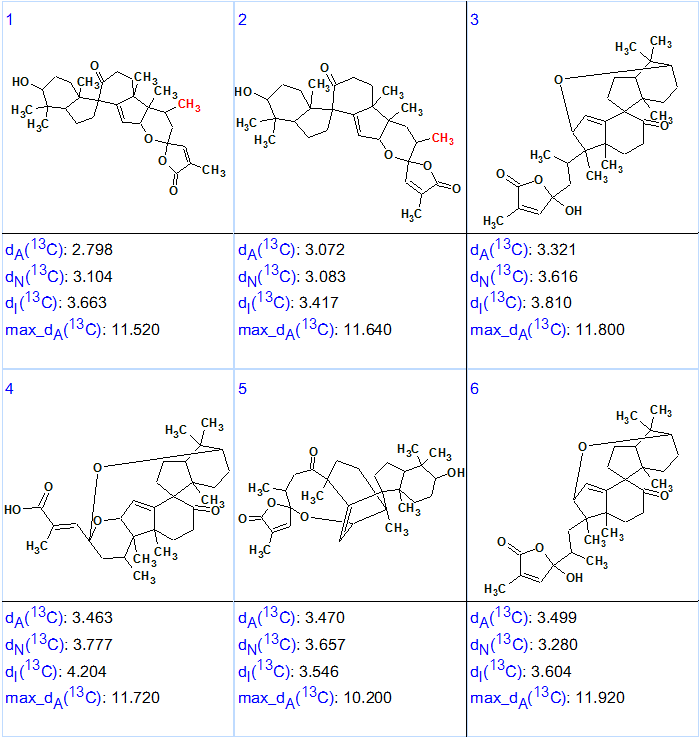
Figure 2. Six top structures of the ranked output structural file for Spirochensilide A.
The top-ranked structure of Figure 2 is identical to structure of Spirochensilide A (1). This same structure, with 13C chemical shift assignments made by the program, is shown below:
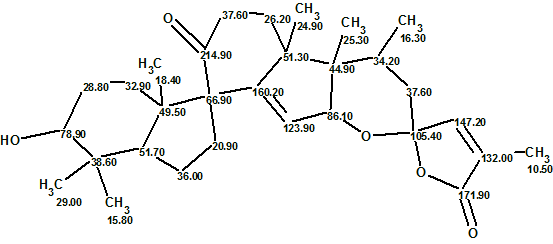
We see from Figure 2 that the values of average deviations calculated for structure #1 are relatively large and do not differ significantly from the deviations calculated for structure #2. Analysis of the 13C chemical shift prediction protocols showed that largest errors were associated with δC calculation for the atoms CH2 36.00, CH2 20.90 and C 160.20 of this unusual structure. Note that δC=160.2 ppm is more typical for oxygenated sp2-hybridized carbons. However, when carbon C 160.2 was labeled with sp2/ob on the MCD, the program produced four exotic structures with deviation values 6.5–8.5 ppm in 8s (Figure 3).
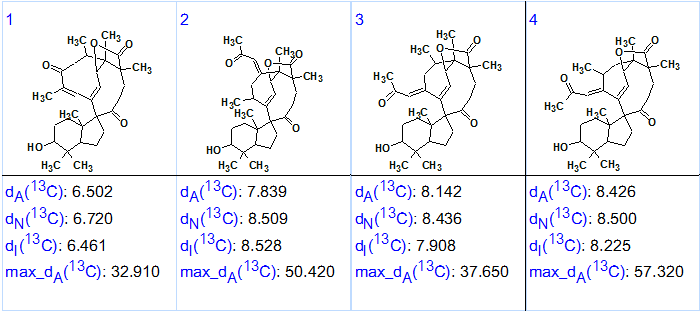
Figure 3. Ranked output structural file for Spirochensilide A obtained as a result of checking validity of “C 160.2 sp2/ob” axiom.
This secondary experiment clearly demonstrates how the program allows one to quickly check any alternative theory/axiom [2] and consequently select the most appropriate one.
It is worthwhile to note that there is a single difference between structures #1 and #2 for carbon atom CH3 C 16.3 (marked by red color). The original authors [1] detected a NOESY correlation of this methyl to the CH3 24.9 group. Building a simple 2D model of structure #2 using ACD/3D Viewer allowed us to approximately estimate the distance between these groups in space to be >6 Å. This indicates that the hydrogens of these methyl groups likely cannot exhibit a NOESY correlation, a fact confirmed by the priority ranking of structure #1.
References
- Q.-Q. Zhao, Q.-Y. Song, K. Jiang, G.-D. Li, W.-J. Wei, Y. Li, K. Gao. (2015). Spirochensilides A and B, Two New Rearranged Triterpenoids from Abies chensiensis. Org. Lett. 17:2760–2763.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.


