October 1, 2014
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Strophasterol A
In search for bioactive compounds from the mushroom Stropharia rugosoannulata, Wu et al [1] discovered four novel steroids named strophasterols A, B, C, and D which contained a very unique and unprecedented carbon skeleton. Here we will describe the computerized structure determination of Strophasterol A (1).
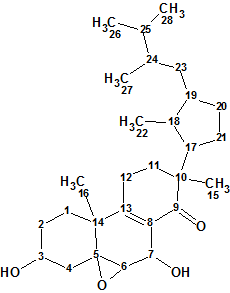
1
Strophasterol A was obtained as white crystals. Its molecular formula was determined to be C28H44O4 by HRESIMS with m/z 467.3100 [M+Na]+ (calculated as C28H44NaO4 467.3137), thus indicating the molecule contained seven degrees of unsaturation. The planar structure of 1 was elucidated by interpretation of the NMR spectra, including DEPT, COSY, HMBC and HMQC data. Confirmation of the planar structure and determination of its absolute and relative configuration were performed by X-ray crystallography analysis of its bis(p-bromo)benzoate derivative. The DEPT experiment indicated the presence of six methyl, eight methylene, and eight methine groups, as well as six quaternary carbon atoms.
The experimental 1H, 13C, HMQC and HMBC data were tabulated in the article [1]. They are presented in Table 1 and their graphical mapping in the form of the Molecular Connectivity Diagram (MCD) is shown in Figure 1.
Table 1: Spectroscopic NMR data (in CDCl3) for
Strophasterol A.
| Label | δC | δC calc | CHn | δH | M(J) | C HMBC |
| C 1 | 29.2 | 30.81 | CH2 | 1.7 | u | C 5, C 14, C 3, C 16 |
| C 1 | 29.2 | 30.81 | CH2 | 1.84 | u | |
| C 2 | 30.4 | 30.93 | CH2 | 1.99 | u | |
| C 2 | 30.4 | 30.93 | CH2 | 1.69 | u | C 16, C 1, C 4, C 3 |
| C 3 | 68.2 | 68.62 | CH | 3.94 | u | |
| C 4 | 38.8 | 38.08 | CH2 | 2.19 | u | |
| C 4 | 38.8 | 38.08 | CH2 | 1.48 | u | C 3, C 6, C 5, C 2, C 14 |
| C 5 | 62.5 | 64.98 | C | |||
| C 6 | 59.4 | 62.1 | CH | 3.21 | u | C 8, C 5, C 7, C 4 |
| C 7 | 63 | 63.82 | CH | 4.84 | u | C 5, C 9, C 8, C 13, C 6 |
| C 8 | 128.1 | 134.38 | C | |||
| C 9 | 205.2 | 205.29 | C | |||
| C 10 | 45.6 | 47.73 | C | |||
| C 11 | 32.1 | 34.96 | CH2 | 2 | u | |
| C 11 | 32.1 | 34.96 | CH2 | 1.65 | u | C 12, C 17, C 9, C 15, C 13, C 10 |
| C 12 | 22.2 | 23.13 | CH2 | 2.19 | u | |
| C 12 | 22.2 | 23.13 | CH2 | 2.39 | u | C 8, C 14, C 10, C 13, C 11 |
| C 13 | 159.5 | 158.56 | C | |||
| C 14 | 39.6 | 41.92 | C | |||
| C 15 | 19 | 21.98 | CH3 | 0.93 | u | C 17, C 10, C 9, C 11 |
| C 16 | 22.3 | 21.06 | CH3 | 1.31 | u | C 1, C 14, C 5, C 13 |
| C 17 | 47.7 | 52.42 | CH | 1.88 | u | C 9, C 15, C 20, C 21, C 22, C 18, C 10 |
| C 18 | 41.5 | 38.01 | CH | 1.25 | u | C 17, C 22, C 19, C 20, C 23 |
| C 19 | 46.4 | 41.96 | CH | 1.36 | u | C 26, C 22, C 28, C 25, C 23 |
| C 20 | 31.8 | 30.89 | CH2 | 1.73 | u | |
| C 20 | 31.8 | 30.89 | CH2 | 1.03 | u | C 21, C 19, C 18, C 17, C 23 |
| C 21 | 26.4 | 27.42 | CH2 | 1.37 | u | |
| C 21 | 26.4 | 27.42 | CH2 | 1.27 | u | C 17, C 20, C 18, C 19, C 10 |
| C 22 | 20.8 | 17.9 | CH3 | 0.97 | u | C 19, C 18, C 17 |
| C 23 | 39.58 | 37.53 | CH2 | 0.86 | u | C 19, C 25, C 20, C 27, C 24, C 18 |
| C 23 | 39.58 | 37.53 | CH2 | 1.44 | u | |
| C 24 | 36.7 | 36.78 | CH | 1.36 | u | |
| C 25 | 30.2 | 32.39 | CH | 1.6 | u | C 23, C 26, C 28, C 27 |
| C 26 | 16.4 | 18.67 | CH3 | 0.72 | u | C 25, C 24, C 28 |
| C 27 | 15.6 | 15.84 | CH3 | 0.74 | u | C 25, C 24, C 23 |
| C 28 | 20.7 | 20.87 | CH3 | 0.83 | u | C 25, C 26, C 24 |
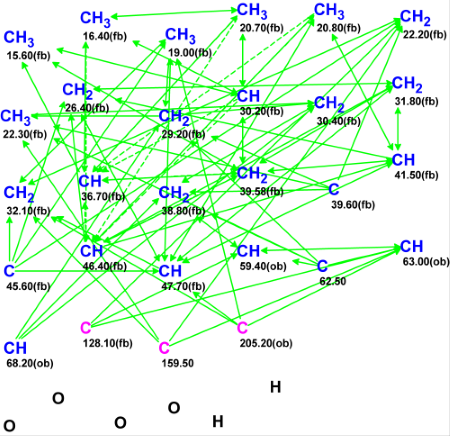
Figure 1. The Molecular Connectivity Diagram (MCD) as originally
generated for Strophasterol A.
MCD overview. Two specific attributes for the MCD are: a) there are many sp3-hybridized carbon atoms which are labelled by the program as “fb” (forbidden, oxygens cannot be connected to these atoms); and b) the connectivity net is dense, i.e. the number of HMBC correlations (68) is high. Under such conditions, we can expect that structure generation can be successfully performed without any MCD edits though some of them are obvious (for example, carbons C 63.00 (4.84) and C 68.2 (3.94) definitely have an oxygen atom as a neighbor).
MCD checking revealed the presence of contradictions in the HMBC data. Therefore, Fuzzy Structure Generation was initiated in the mode “Determine Options Automatically” with the results: k=152→23→6, tg= 1s; 1 of the 68 HMBC connectivities was extended during the structure generation process, and 5 of the 68 possible connectivity combinations were used during generation. The three top structures of the ranked output file are presented in Figure 2.
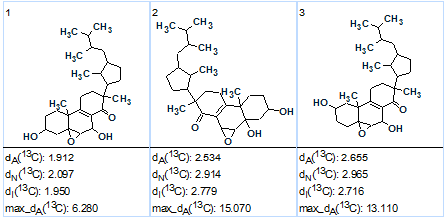
Figure 2. Strophasterol A.The three top structures of the ranked output file.
It is obvious that structure #1 is identical to structure 1 of Strophasterol A determined in the original work [1]. The closest competing structures are similar to structure #1, but they are reliably ruled out by the program from the set of candidate structures. Structure 2 shows the 13C chemical shift assignment carried out automatically during structure generation.
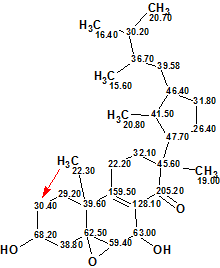
2
Thus a molecule containing an unprecedented skeleton was automatically and instantly identified without the utilization of X-ray analysis or user intervention.
References
- Wu J, Tokuyama S, Nagai K, Yasuda N, Noguchi K, Matsumoto T, Hirai H, Kawagishi H. (2012). Strophasterols A to D with an unprecedented steroid skeleton: from the mushroom Stropharia rugosoannulata. Angew Chem Int Ed Engl, 51(43):10820–10822.


