November 1, 2014
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Strynuxline A
Strychnos nux-vomica is a moderate-sized tree of the family Loganiaceae found in southern Asian countries. The dried ripe seeds of this tree have been applied as a traditional folk medicine in China for the treatment of tumors, rheumatic arthritis, swelling pain, trauma, bone fractures, etc. Strychnine and brucine are major components, which are also mainly responsible for most of the pharmacological properties such as antitumor, cytoprotective, and antitussive activities. Studies aimed at the discovery of trace alkaloids in S. nux-vomica led Fu and co-workers [1] to the isolation of Strynuxlines A and B, two novel alkaloids possessing an unprecedented skeleton with a 6/5/9/6/7/6 hexacyclic ring system. This was the first report of the cleavage of the C-3 – C-7 bond in strychnan-type alkaloids. In addition to Strynuxlines A and B, 16 known alkaloids were isolated and identified in this work.
Here we will describe how Strynuxlines A (1) could be identified with the assistance of a CASE-based approach using ACD/Structure Elucidator Suite.
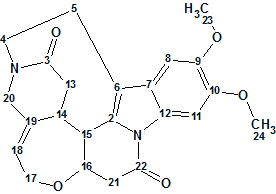
1
Strynuxline A (1) was obtained as a white, amorphous solid. Its molecular formula was determined to be C23H24N2O5, by HREIMS (m/z 408.1673 [M]+; calculated 408.1685), with an index of hydrogen deficiency of 13. The 13C, 1H and HSQC spectroscopic data, as well as key HMBC and COSY correlations presented in the article [1] graphically are given in Table 1.
Table 1. Spectroscopic NMR data for Strynuxline A.
| Label | dC | δCcalc | CHn | δH | M(J) | COSY | C HMBC |
| C 2 | 130.1 | 133.47 | C | ||||
| C 3 | 174.9 | 171.59 | C | ||||
| C 5 | 49.3 | 47.49 | CH2 | 3.17 | u | 2.8 | C 7, C 3, C 21 |
| C 5 | 49.3 | 47.49 | CH2 | 4.57 | u | ||
| C 6 | 25.7 | 22.64 | CH2 | 3.14 | u | ||
| C 6 | 25.7 | 22.64 | CH2 | 2.8 | u | 3.17 | C 7, C 8 |
| C 7 | 117.7 | 113.15 | C | ||||
| C 8 | 124.7 | 123.54 | C | ||||
| C 9 | 100.4 | 100.28 | CH | 6.78 | s | C 7, C 11, C 8 | |
| C 10 | 146.9 | 146.2 | C | ||||
| C 11 | 148.1 | 146.88 | C | ||||
| C 12 | 99.7 | 98.38 | CH | 7.88 | s | C 8, C 10 | |
| C 13 | 129.5 | 131.32 | C | ||||
| C 14 | 39.5 | 35.25 | CH2 | 2.86 | dd(15.1, 6.3) | ||
| C 14 | 39.5 | 35.25 | CH2 | 2.48 | d(15.1) | 3.51 | |
| C 15 | 41.5 | 35.44 | CH | 3.51 | u | 3.29, 2.48 | C 19, C 2, C 3 |
| C 16 | 48.7 | 49.95 | CH | 3.29 | u | 3.51, 4.13 | |
| C 17 | 83.7 | 76.79 | CH | 4.13 | u | 2.77, 3.29 | C 7, C 18, C 15, C 24, C 16 |
| C 18 | 68 | 65.19 | CH2 | 4.10 | d(13.8) | 6.01 | C 20 |
| C 18 | 68 | 65.19 | CH2 | 4.26 | dd(13.8, 7.0) | ||
| C 19 | 123.3 | 126.68 | CH | 6.01 | u | 4.1 | |
| C 20 | 145.1 | 133.39 | C | ||||
| C 21 | 52.9 | 53.75 | CH2 | 4.19 | u | C 15, C 19, C 3 | |
| C 21 | 52.9 | 53.75 | CH2 | 3.86 | u | ||
| C 23 | 42.3 | 42.3 | CH2 | 2.77 | u | 4.13 | C 24 |
| C 23 | 42.3 | 42.3 | CH2 | 3.15 | u | ||
| C 24 | 168 | 166.35 | C | ||||
| C 25 | 56.3 | 56.34 | CH3 | 3.86 | s | ||
| C 26 | 56.3 | 56.47 | CH3 | 3.86 | s | C 11, C 10 |
Figure 1 shows the initial Molecular Connectivity Diagram created from the NMR spectroscopic data.
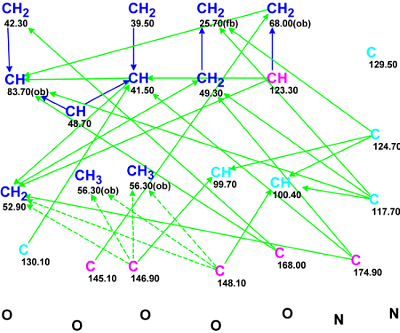
Figure 1. The Molecular Connectivity Diagram for Strynuxline A.
MCD overview. Only four carbon atoms C 56.3 – C 83.7 were fully assigned by the program, while six carbons (marked by a light blue color) were assigned an ambiguous state of hybridization (sp3 or sp2) and the possibility of a heteroatom neighborhood is also undefined. Taking into account the 1H chemical shifts of hydrogens (7.88 and 6.78 ppm) attached to atoms C 99.7 and C 100.4 respectively, as well as the characteristic 13C chemical shifts of the other four “light blue” quaternary carbons, a hybridization state of sp2 can be assigned to all of them. Two absorption bands were observed at 1670 and
1640 cm-1 in the IR spectrum. Therefore it is possible to suggest that the carbons at C 168.0 and C 174.9 are connected to oxygen atoms with double bonds. Two methyl groups at C 56.3 can be safely connected to oxygen atoms. With the presence of two nitrogen atoms in the molecule, it would be risky to assign the additional labels “ob” or “fb” (obligatory or forbidden) to carbon atoms displayed on the MCD. The numbers of hydrogen atoms present in the first sphere around eight carbon atoms were set on the MCD in agreement with the 1H signal multiplicities shown in Table 1. The MCD, edited in accordance with given above considerations, is shown in Figure 2.
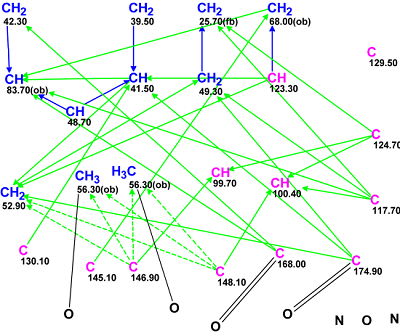
Figure 2. The edited Molecular Connectivity Diagram for Strynuxline A.
MCD checking with the program produced a message indicating that no contradictions were detected in the 2D NMR data. Strict structure generation was initiated which gave the following result: k = 132→94→77, tg = 0.27 s. The three top structures of the ranked output file are presented in Figure 3.
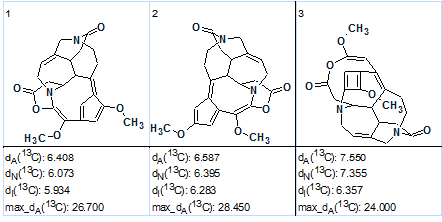
Figure 3. The three top structures of the ranked output file produced by strict structure generation.
The large values of the average deviations, as well as the “exotic” structure skeletons, suggest that the solution is wrong and the next most rationale step is an attempt at Fuzzy Structure Generation. With options m=1, a=16 set, Fuzzy Structure Generation was completed with the results: k = 5142 → 3411→ 1883, tg= 25 s, where 30 out of 30 possible connectivity combinations were used during the generation process. The first three structures of the ranked structural file are shown in Figure 4.
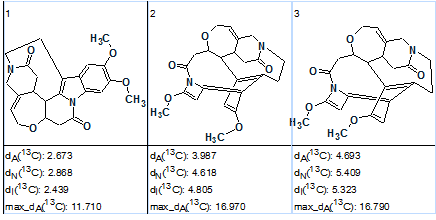
Figure 4. The three top structures of the ranked output file produced by the Fuzzy Structure Generation.
The first-ranked structure coincides with structure 1 originally derived by the authors. [1] The 13C chemical shift assignment is shown on structure 2 (a red arrow shows the detected nonstandard HMBC correlation):
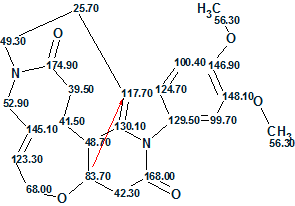
2
This example demonstrates that a complex and unique structure, despite a very unusual skeleton, can be quickly and unambiguously determined along with its chemical shift assignments by Structure Elucidator Suite.
References
- Fu Y, Zhang Y, He H, Hou L, Di Y, Li S, Luo X, Hao X. (2012). Strynuxlines A and B, alkaloids with an unprecedented carbon skeleton from Strychnos nux-vomica. J Nat Prod 75 (11), 1987–1990. doi:10.1021/np300339r


