March 1, 2012
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Tasiamide B
The Initial Challenge
In 2003, Philip Williams and co-workers1 isolated a new cytotoxic peptide, Tasiamide B, from the marine cyanobacterium Symploca sp, which contained the unusual amino acid-derived residue 4-amino-3-hydroxy-5-phenylpentanoic acid (AHPPA). The structure of the peptide was determined through a combination of 2D NMR experiments and HPLC analysis of degradation products. The initial experimental data was then submitted to the Structure Elucidator Challenge to determine if the software could propose the same structure.
The Evidence
- The molecular formula of Tasiamide B was determined to be C50H74N8O12
- The following data were acquired:
- 2D 1H-1H COSY (33 correlations), 1H-13C HMBC (59 correlations)
The CASE (Computer Assisted Structure Elucidation) Investigation
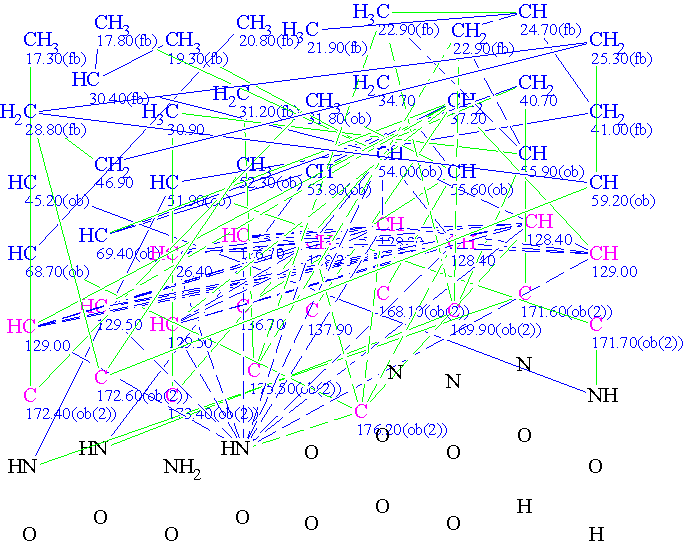
The Molecular Connectivity Diagram produced by Structure Elucidator
-
- Initially, after 15 hours Structure Elucidator was unable to produce a structure consistent with the experimental data, which included 9 carbon atoms with sp2 hybridization according to the authors’ findings.
- Since the 13C NMR showed only 46 signals instead of 50, it was deduced that molecule contained fragments exhibiting local symmetry. The authors deduced the presence of two monosubstituted phenyl rings.
- Two 1-AR (aromatic) fragments were introduced as User Fragments (UF) in the software, which promptly solved the structure in Fragment Mode in 23 seconds.
- Structure 1 ranked in the first position based on the lower dA value; the average deviation (dA) is a comparison between the experimental and predicted 13C chemical shifts.
| Structure #1 | Structure #2 | Structure #3 |
| dA = 1.227 | dA = 1.781 | dA = 1.843 |
| dF = 2.113 | dF = 2.253 | dF = 2.573 |
| dH = 0.188 | dH = 0.206 | dH = 0.242 |
| dΣ = 3.122 | dΣ = 3.841 | dΣ = 4.261 |
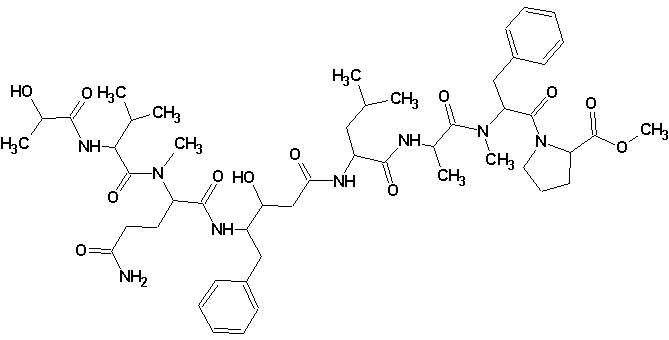
Structure 1
- Structure 1 matches the structure published by the original authors who solved it by traditional means.
Conclusions
The addition of two 1-AR fragments helped to reduce the number of ambiguous connectivities and ultimately reduce the structures generation time. The ambiguity arises from the coincidental chemical shifts of hydrogen atoms in the aromatic region.
The problem demonstrates the benefits of introducing User Fragments which are easily revealed in the experimental data during the preliminary evaluation.
- dF—the average deviation between the experimentally observed carbon chemical shifts and the carbon chemical shifts predicted using fast (increment based) prediction methods
- dH—the average deviation between the experimentally observed proton chemical shifts and the predicted proton chemical shifts using accurate (HOSE code based) prediction methods
- dΣ = dA + 20-dH—generalized match factor that takes into account deviations calculated by the accurate prediction methods both for 13C and 1H NMR spectra
References
- P.G. Williams, W.Y. Yoshida, R.E. Moore, V.J. Paul J. Nat. Prod., 66:1006–1009, 2003.


