November 1, 2015
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Tronoharine
The genus Tabernaemontana (Apocynaceae) incorporates a large number of species, which are distributed over the tropical and subtropical regions of the world. Plants of this genus are prolific producers of alkaloids, in particular indole and bisindole alkaloids, and are well known to elaborate alkaloids with structurally novel molecular skeletons and useful biological activities. A number of the Malaysian Tabernaemontana have been the subject of thorough chemical studies, with the discovery of many new and biologically active alkaloids. Sim et al [1] reported the isolation, structure determination, and biological activity of seven new alkaloids as well as the structure revision of the known hexacyclic alkaloid Tronoharine.
Tronoharine was isolated earlier in a small amount by the same group as a minor alkaloid from another sample of T. corymbosa collected at a different location [2]. The proposed structure 1 (molecular formula C21H24N2O2) was based on analysis of the MS and NMR data.
1
In the more recent work [1], Tronoharine was obtained in a more significant amount, which has enabled the authors to reevaluate the previously proposed structure. The spectroscopic data of the revised structure 2 were virtually in complete agreement with the previous data, except for the HMBC data where two key three-bond correlations (H-9 to C-7, and H-6 to C-8) were observed in the last instance. Those were not detected in the previously obtained HMBC spectrum due to the limitations imposed by the small quantity of the sample. Correlations H-9 to C-7 and H-6 to C-8 are shown on structures 1 and 2.
2
We decided to investigate whether ACD/Structure Elucidator Suite would be capable to determine the correct structure 2 under the original conditions, where only limited HMBC data (correlations H-9 to C-7 and H-6 to C-8 are omitted) were available. The original NMR spectroscopic data were input into the program (Table 1).
Table 1. NMR spectroscopic data for Tronohorin.
| C Label | δC | δC Calc | XHn | δH | COSY | C HMBC |
| C 2 | 142 | 138.95 | C | |||
| C 3 | 45.8 | 44.88 | CH2 | 2.99 | ||
| C 3 | 45.8 | 44.88 | CH2 | 2.42 | 1.75 | C 15 |
| C 5 | 53.8 | 53.79 | CH2 | 3.01 | ||
| C 5 | 53.8 | 53.79 | CH2 | 2.84 | 1.7 | C 3 |
| C 6 | 43.2 | 40.2 | CH2 | 1.7 | 2.84 | C 2, C 8* |
| C 6 | 43.2 | 40.2 | CH2 | 2.69 | ||
| C 7 | 50.2 | 52.14 | C | |||
| C 8 | 137.9 | 134.86 | C | |||
| C 9 | 120.5 | 121.96 | CH | 7.2 | 7.12 | C 13, C 7* |
| C 10 | 125.3 | 123.79 | CH | 7.12 | 7.24, 7.20 | |
| C 11 | 127.5 | 128.1 | CH | 7.24 | 7.12, 8.07 | |
| C 12 | 115.5 | 111.88 | CH | 8.07 | 7.24 | C 8 |
| C 13 | 138.7 | 140.26 | C | |||
| C 14 | 27.2 | 28.46 | CH2 | 1.75 | 2.32, 2.42 | |
| C 14 | 27.2 | 28.46 | CH2 | 1.85 | ||
| C 15 | 35.3 | 38.62 | CH | 2.32 | 1.75, 1.98 | |
| C 16 | 113.2 | 117.14 | C | |||
| C 17 | 34.6 | 36.62 | CH2 | 2.73 | ||
| C 17 | 34.6 | 36.62 | CH2 | 2.18 | 4.49 | C 2, C 15, C 22 |
| C 18 | 11.7 | 11.44 | CH3 | 0.7 | 0.8 | |
| C 19 | 23.7 | 23.84 | CH2 | 0.8 | 1.98, 0.70 | |
| C 20 | 42.5 | 41.43 | CH | 1.98 | 0.80, 3.73, 2.32 | C 7, C 16 |
| C 21 | 65.8 | 67.56 | CH | 3.73 | 1.98 | C 2, C 8, C 15 |
| C 22 | 169 | 170.4 | C | |||
| C 23 | 67.7 | 68.39 | CH | 4.49 | 2.18 |
* Correlations from the secondary work [2] which were not introduced for the first program run.
From the data in Table 1, a Molecular Connectivity Diagram (MCD) was created by the software:
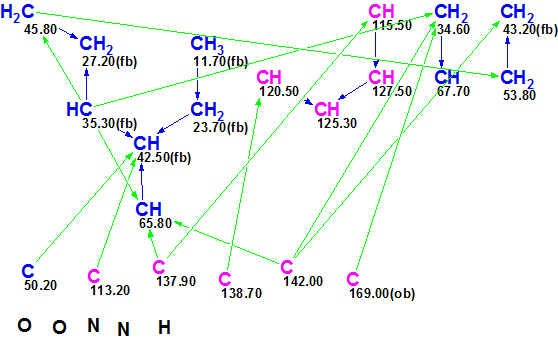
Figure 1. Molecular Connectivity Diagram (MCD) for Tronohorin.
MCD checking showed that the 2D NMR data are consistent and, therefore, a Strict Structure Generation followed with 13C chemical shift prediction and structure filtering at the threshold dI=5 ppm was performed, which gave the following results: k=283936 → 257 → 171, tg=14m. The top 12 structures of the ranked output structural file are presented in Figure 2.
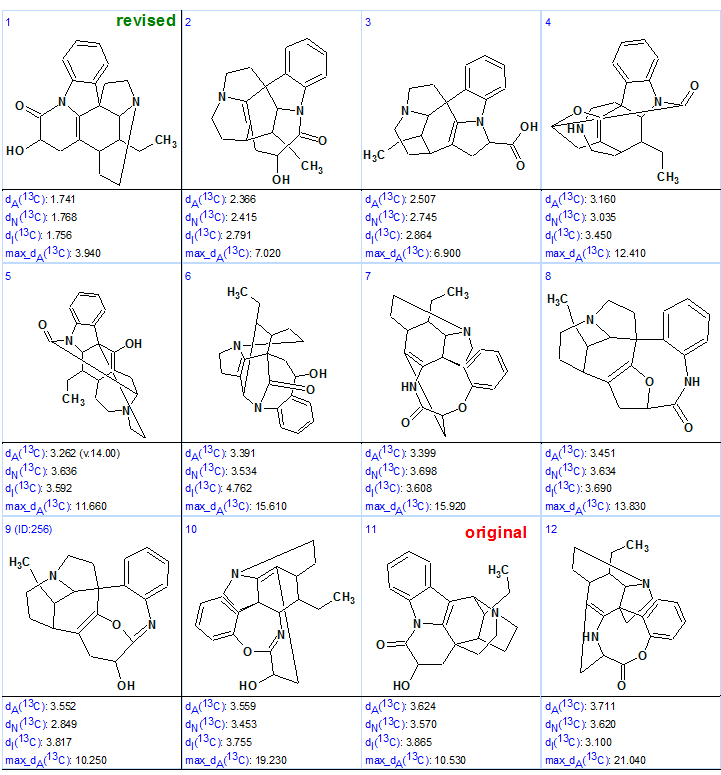
Figure 2. Top 12 structures of the ranked output structural file for Tronohorin.
Figure 2 shows that the correct structure 2 of Tronohorin was generated by Structure Elucidator Suite from the original limited NMR data, and the ranking procedure distinguished it as the best structure. At the same time, the original structure 1 was also generated, but placed on the 11 position by the ranking procedure. The revised structure with automatic 13C chemical shift assignment is shown below:
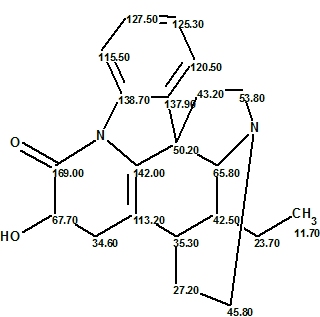
This example demonstrates the ability of ACD/Structure Elucidator Suite to quickly determine the correct structure of a novel compound, even when only limited/incomplete NMR data is available.
References
- D. S.-Y. Sim, K.-W. Chong, C.-E. Nge, Y.-Y. Low, K.-S. Sim, T.-S. Kam. (2014). Cytotoxic Vobasine, Tacaman, and Corynanthe-Tryptamine Bisindole Alkaloids from Tabernaemontana and Structure Revision of Tronoharine. J. Nat. Prod. 77:2504–2512
- T. S. Kam, K. M. Sim, T. M. Lim. (1999). Tetrahedron Lett. 40:5409–5412.


